当前位置:
X-MOL 学术
›
Mol. Microbiol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
De novo genome assembly of Candida glabrata reveals cell wall protein complement and structure of dispersed tandem repeat arrays.
Molecular Microbiology ( IF 2.6 ) Pub Date : 2020-02-18 , DOI: 10.1111/mmi.14488 Zhuwei Xu 1 , Brian Green 1 , Nicole Benoit 1 , Michael Schatz 2 , Sarah Wheelan 3 , Brendan Cormack 1
Molecular Microbiology ( IF 2.6 ) Pub Date : 2020-02-18 , DOI: 10.1111/mmi.14488 Zhuwei Xu 1 , Brian Green 1 , Nicole Benoit 1 , Michael Schatz 2 , Sarah Wheelan 3 , Brendan Cormack 1
Affiliation
|
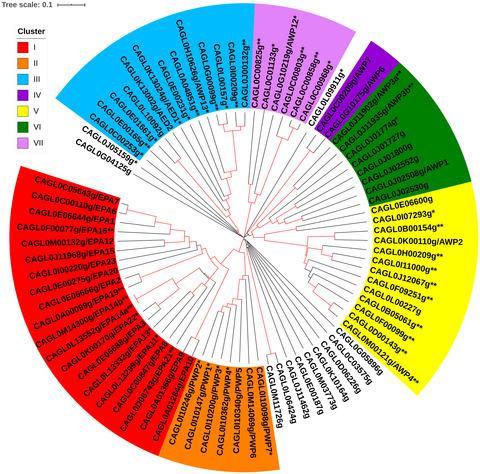
Candida glabrata is an opportunistic pathogen in humans, responsible for approximately 20% of disseminated candidiasis. Candida glabrata's ability to adhere to host tissue is mediated by GPI‐anchored cell wall proteins (GPI‐CWPs); the corresponding genes contain long tandem repeat regions. These repeat regions resulted in assembly errors in the reference genome. Here, we performed a de novo assembly of the C. glabrata type strain CBS138 using long single‐molecule real‐time reads, with short read sequences (Illumina) for refinement, and constructed telomere‐to‐telomere assemblies of all 13 chromosomes. Our assembly has excellent agreement overall with the current reference genome, but we made substantial corrections within tandem repeat regions. Specifically, we removed 62 genes of which 45 were scrambled due to misassembly in the reference. We annotated 31 novel ORFs of which 24 ORFs are GPI‐CWPs. In addition, we corrected the tandem repeat structure of an additional 21 genes. Our corrections to the genome were substantial, with the length of new genes and tandem repeat corrections amounting to approximately 3.8% of the ORFeome length. As most corrections were within the coding regions of GPI‐CWP genes, our genome assembly establishes a high‐quality reference set of genes and repeat structures for the functional analysis of these cell surface proteins.
中文翻译:

从头念珠菌的基因组大会揭示了细胞壁蛋白补体和分散的串联重复阵列的结构。
光滑念珠菌是人类的机会病原体,约占散发性念珠菌病的20%。光滑念珠菌粘附宿主组织的能力是由GPI锚定的细胞壁蛋白(GPI-CWPs)介导的。相应的基因包含长串联重复序列区域。这些重复区域导致参考基因组中的装配错误。在这里,我们进行了C. glabrata的从头组装使用长单分子实时读取,短读取序列(Illumina)进行精制的CBS138型菌株,并构建了所有13条染色体的端粒到端粒装配体。我们的程序集与当前的参考基因组总体上具有极好的一致性,但是我们在串联重复区域内进行了实质性的更正。具体来说,我们删除了62个基因,其中45个由于参比中的错误组装而混乱。我们注释了31个新颖的ORF,其中24个ORF是GPI-CWP。此外,我们更正了另外21个基因的串联重复结构。我们对基因组的校正非常重要,新基因的长度和串联重复校正的长度约为ORFeome长度的3.8%。由于大多数校正均在GPI-CWP基因的编码区域内,
更新日期:2020-02-18
中文翻译:

从头念珠菌的基因组大会揭示了细胞壁蛋白补体和分散的串联重复阵列的结构。
光滑念珠菌是人类的机会病原体,约占散发性念珠菌病的20%。光滑念珠菌粘附宿主组织的能力是由GPI锚定的细胞壁蛋白(GPI-CWPs)介导的。相应的基因包含长串联重复序列区域。这些重复区域导致参考基因组中的装配错误。在这里,我们进行了C. glabrata的从头组装使用长单分子实时读取,短读取序列(Illumina)进行精制的CBS138型菌株,并构建了所有13条染色体的端粒到端粒装配体。我们的程序集与当前的参考基因组总体上具有极好的一致性,但是我们在串联重复区域内进行了实质性的更正。具体来说,我们删除了62个基因,其中45个由于参比中的错误组装而混乱。我们注释了31个新颖的ORF,其中24个ORF是GPI-CWP。此外,我们更正了另外21个基因的串联重复结构。我们对基因组的校正非常重要,新基因的长度和串联重复校正的长度约为ORFeome长度的3.8%。由于大多数校正均在GPI-CWP基因的编码区域内,











































 京公网安备 11010802027423号
京公网安备 11010802027423号