当前位置:
X-MOL 学术
›
Hum. Mutat.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Single-molecule detection of cancer mutations using a novel PCR-LDR-qPCR assay.
Human Mutation ( IF 3.9 ) Pub Date : 2020-02-17 , DOI: 10.1002/humu.23987 Cristian Ruiz 1, 2 , Jianmin Huang 1 , Sarah F Giardina 1 , Philip B Feinberg 1 , Aashiq H Mirza 1 , Manny D Bacolod 1 , Steven A Soper 3 , Francis Barany 1
Human Mutation ( IF 3.9 ) Pub Date : 2020-02-17 , DOI: 10.1002/humu.23987 Cristian Ruiz 1, 2 , Jianmin Huang 1 , Sarah F Giardina 1 , Philip B Feinberg 1 , Aashiq H Mirza 1 , Manny D Bacolod 1 , Steven A Soper 3 , Francis Barany 1
Affiliation
|
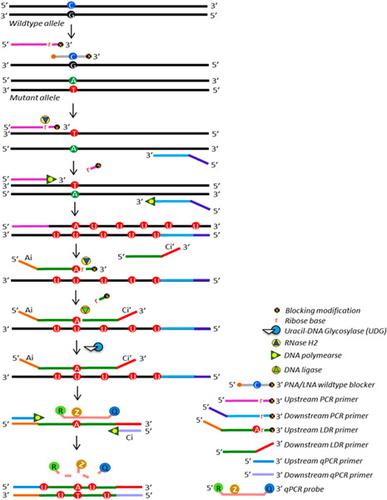
Detection of low-abundance mutations in cell-free DNA is being used to identify early cancer and early cancer recurrence. Here, we report a new PCR-LDR-qPCR assay capable of detecting point mutations at a single-molecule resolution in the presence of an excess of wild-type DNA. Major features of the assay include selective amplification and detection of mutant DNA employing multiple nested primer-binding regions as well as wild-type sequence blocking oligonucleotides, prevention of carryover contamination, spatial sample dilution, and detection of multiple mutations in the same position. Our method was tested to interrogate the following common cancer somatic mutations: BRAF:c.1799T>A (p.Val600Glu), TP53:c.743G>A (p.Arg248Gln), KRAS:c.35G>C (p.Gly12Ala), KRAS:c.35G>T (p.Gly12Val), KRAS:c.35G>A (p.Gly12Asp), KRAS:c.34G>T (p.Gly12Cys), and KRAS:c.34G>A (p.Gly12Ser). The single-well version of the assay detected 2-5 copies of these mutations, when diluted with 10,000 genome equivalents (GE) of wild-type human genomic DNA (hgDNA) from buffy coat. A 12-well (pixel) version of the assay was capable of single-molecule detection of the aforementioned mutations at TP53, BRAF, and KRAS (specifically p.Gly12Val and p.Gly12Cys), mixed with 1,000-2,250 GE of wild-type hgDNA from plasma or buffy coat. The assay described herein is highly sensitive, specific, and robust, and potentially useful in liquid biopsies.
中文翻译:

使用新型PCR-LDR-qPCR分析法对癌症突变进行单分子检测。
检测无细胞DNA中的低丰度突变可用于识别早期癌症和早期癌症复发。在这里,我们报告了一种新的PCR-LDR-qPCR分析方法,该方法能够在存在过量野生型DNA的情况下以单分子分辨率检测点突变。该测定法的主要特征包括使用多个嵌套引物结合区以及野生型序列阻断性寡核苷酸选择性扩增和检测突变型DNA,防止残留污染,空间样品稀释以及在同一位置检测多个突变。我们的方法经过测试可查询以下常见的癌症体细胞突变:BRAF:c.1799T> A(p.Val600Glu),TP53:c.743G> A(p.Arg248Gln),KRAS:c.35G> C(p.Gly12Ala ),KRAS:c.35G> T(p.Gly12Val),KRAS:c.35G> A(p.Gly12Asp),KRAS:c.34G> T(p.Gly12Val)Gly12Cys)和KRAS:c.34G> A(p.Gly12Ser)。当用来自血沉棕黄层的野生型人基因组DNA(hgDNA)的10,000个基因组当量(GE)稀释时,该测定的单孔形式检测到2-5个拷贝的这些突变。该分析的12孔(像素)版本能够单分子检测TP53,BRAF和KRAS(特别是p.Gly12Val和p.Gly12Cys)上的上述突变,并与1,000-2,250 GE的野生型混合来自血浆或血沉棕黄层的hgDNA。本文所述的测定是高度灵敏的,特异性的和鲁棒的,并且在液体活检中可能有用。该分析的12孔(像素)版本能够单分子检测TP53,BRAF和KRAS(特别是p.Gly12Val和p.Gly12Cys)上的上述突变,并与1,000-2,250 GE的野生型混合来自血浆或血沉棕黄层的hgDNA。本文所述的测定是高度灵敏的,特异性的和鲁棒的,并且在液体活检中可能有用。该分析的12孔(像素)版本能够单分子检测TP53,BRAF和KRAS(特别是p.Gly12Val和p.Gly12Cys)上的上述突变,并与1,000-2,250 GE的野生型混合来自血浆或血沉棕黄层的hgDNA。本文所述的测定是高度灵敏的,特异性的和鲁棒的,并且在液体活检中可能有用。
更新日期:2020-02-17
中文翻译:

使用新型PCR-LDR-qPCR分析法对癌症突变进行单分子检测。
检测无细胞DNA中的低丰度突变可用于识别早期癌症和早期癌症复发。在这里,我们报告了一种新的PCR-LDR-qPCR分析方法,该方法能够在存在过量野生型DNA的情况下以单分子分辨率检测点突变。该测定法的主要特征包括使用多个嵌套引物结合区以及野生型序列阻断性寡核苷酸选择性扩增和检测突变型DNA,防止残留污染,空间样品稀释以及在同一位置检测多个突变。我们的方法经过测试可查询以下常见的癌症体细胞突变:BRAF:c.1799T> A(p.Val600Glu),TP53:c.743G> A(p.Arg248Gln),KRAS:c.35G> C(p.Gly12Ala ),KRAS:c.35G> T(p.Gly12Val),KRAS:c.35G> A(p.Gly12Asp),KRAS:c.34G> T(p.Gly12Val)Gly12Cys)和KRAS:c.34G> A(p.Gly12Ser)。当用来自血沉棕黄层的野生型人基因组DNA(hgDNA)的10,000个基因组当量(GE)稀释时,该测定的单孔形式检测到2-5个拷贝的这些突变。该分析的12孔(像素)版本能够单分子检测TP53,BRAF和KRAS(特别是p.Gly12Val和p.Gly12Cys)上的上述突变,并与1,000-2,250 GE的野生型混合来自血浆或血沉棕黄层的hgDNA。本文所述的测定是高度灵敏的,特异性的和鲁棒的,并且在液体活检中可能有用。该分析的12孔(像素)版本能够单分子检测TP53,BRAF和KRAS(特别是p.Gly12Val和p.Gly12Cys)上的上述突变,并与1,000-2,250 GE的野生型混合来自血浆或血沉棕黄层的hgDNA。本文所述的测定是高度灵敏的,特异性的和鲁棒的,并且在液体活检中可能有用。该分析的12孔(像素)版本能够单分子检测TP53,BRAF和KRAS(特别是p.Gly12Val和p.Gly12Cys)上的上述突变,并与1,000-2,250 GE的野生型混合来自血浆或血沉棕黄层的hgDNA。本文所述的测定是高度灵敏的,特异性的和鲁棒的,并且在液体活检中可能有用。



























 京公网安备 11010802027423号
京公网安备 11010802027423号