Computational and Structural Biotechnology Journal ( IF 4.4 ) Pub Date : 2020-01-18 , DOI: 10.1016/j.csbj.2020.01.001 Stylianos Mavropoulos Papoudas 1 , Nikolaos Papanikolaou 1 , Christoforos Nikolaou 1, 2
|
Genes in linear proximity often share regulatory inputs, expression and evolutionary patterns, even in complex eukaryote genomes with extensive intergenic sequences. Gene regulation, on the other hand, is effected through the co-ordinated activation (or suppression) of genes participating in common biological pathways, which are often transcribed from distant loci. Existing approaches for the study of gene expression focus on the functional aspect, taking positional constraints into account only marginally.
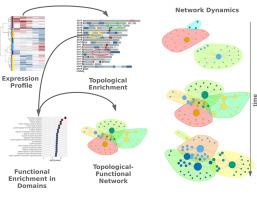
In this work we propose a novel concept for the study of gene expression, through the combination of topological and functional information into bipartite networks. Starting from genome-wide expression profiles, we define extended chromosomal regions with consistent patterns of differential gene expression and then associate these domains with enriched functional pathways. By analyzing the resulting networks in terms of size, connectivity and modularity we can draw conclusions on the way genome organization may underlie the gene regulation program.
Implementation of this approach in a detailed RNASeq profiling of sustained Tnf stimulation of mouse synovial fibroblasts, allowed us to identify unexpected regulatory changes taking place in the cells after 24 h of stimulation. Bipartite network analysis suggests that the cytokine response set by Tnf, progresses through two distinct transitions. An early generalization of the inflammatory response, that is followed by a late shutdown of immune-related functions and the redistribution of expression to developmental and cell adhesion pathways and distinct chromosomal regions.
We show that the incorporation of topological information may provide additional insights in the complex propagation of Tnf activation.
中文翻译:

使用拓扑功能网络监控时间和空间上长时间的Tnf刺激。
线性接近的基因通常共享调节输入,表达和进化模式,即使在具有广泛基因间序列的复杂真核生物基因组中也是如此。另一方面,基因调控是通过参与共同生物学途径的基因的协调激活(或抑制)来实现的,这些生物学途径通常是从远距离基因座转录而来的。研究基因表达的现有方法集中于功能方面,仅略微考虑位置限制。
在这项工作中,我们提出了一个新的概念,通过将拓扑和功能信息组合成二分网络来研究基因表达。从全基因组表达谱开始,我们定义具有一致的差异基因表达模式的扩展染色体区域,然后将这些结构域与丰富的功能途径相关联。通过从大小,连通性和模块性方面分析所得的网络,我们可以得出有关基因组组织可能构成基因调控程序基础的结论。
在小鼠滑膜成纤维细胞持续Tnf刺激的详细RNASeq分析中实施此方法,使我们能够确定刺激24小时后细胞中发生的意外调节变化。双向网络分析表明,由Tnf设定的细胞因子反应通过两个不同的转变进行。炎症反应的早期概括,其后是免疫相关功能的晚期关闭,以及表达向发育和细胞粘附途径以及独特的染色体区域的重新分布。
我们表明,拓扑信息的合并可以提供Tnf激活的复杂传播中的其他见解。










































 京公网安备 11010802027423号
京公网安备 11010802027423号