Computational and Structural Biotechnology Journal ( IF 4.4 ) Pub Date : 2019-12-26 , DOI: 10.1016/j.csbj.2019.12.005 Xiaodi Yang 1 , Shiping Yang 2 , Qinmengge Li 3 , Stefan Wuchty 4, 5, 6, 7 , Ziding Zhang 1
|
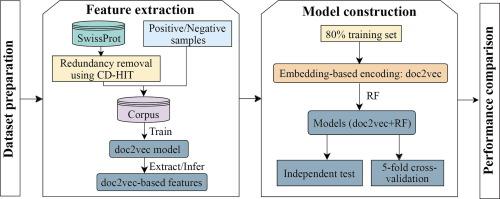
The identification of human-virus protein-protein interactions (PPIs) is an essential and challenging research topic, potentially providing a mechanistic understanding of viral infection. Given that the experimental determination of human-virus PPIs is time-consuming and labor-intensive, computational methods are playing an important role in providing testable hypotheses, complementing the determination of large-scale interactome between species. In this work, we applied an unsupervised sequence embedding technique (doc2vec) to represent protein sequences as rich feature vectors of low dimensionality. Training a Random Forest (RF) classifier through a training dataset that covers known PPIs between human and all viruses, we obtained excellent predictive accuracy outperforming various combinations of machine learning algorithms and commonly-used sequence encoding schemes. Rigorous comparison with three existing human-virus PPI prediction methods, our proposed computational framework further provided very competitive and promising performance, suggesting that the doc2vec encoding scheme effectively captures context information of protein sequences, pertaining to corresponding protein-protein interactions. Our approach is freely accessible through our web server as part of our host-pathogen PPI prediction platform (http://zzdlab.com/InterSPPI/). Taken together, we hope the current work not only contributes a useful predictor to accelerate the exploration of human-virus PPIs, but also provides some meaningful insights into human-virus relationships.
中文翻译:

通过基于序列嵌入的机器学习方法预测人类-病毒蛋白质-蛋白质相互作用。
人-病毒蛋白质-蛋白质相互作用(PPI)的鉴定是一个重要且具有挑战性的研究课题,有可能提供对病毒感染机制的理解。鉴于人类病毒 PPI 的实验测定既耗时又费力,计算方法在提供可检验的假设、补充物种间大规模相互作用组的测定方面发挥着重要作用。在这项工作中,我们应用无监督序列嵌入技术(doc2vec)将蛋白质序列表示为低维的丰富特征向量。通过覆盖人类和所有病毒之间已知 PPI 的训练数据集来训练随机森林 (RF) 分类器,我们获得了出色的预测精度,优于机器学习算法和常用序列编码方案的各种组合。与三种现有的人类病毒 PPI 预测方法进行严格比较,我们提出的计算框架进一步提供了非常有竞争力和有前途的性能,表明 doc2vec 编码方案有效地捕获了与相应的蛋白质-蛋白质相互作用有关的蛋白质序列的上下文信息。我们的方法可以通过我们的网络服务器免费访问,作为我们宿主病原体 PPI 预测平台 (http://zzdlab.com/InterSPPI/) 的一部分。总而言之,我们希望当前的工作不仅可以为加速人类-病毒 PPI 的探索提供有用的预测因子,而且还可以为人类-病毒关系提供一些有意义的见解。









































 京公网安备 11010802027423号
京公网安备 11010802027423号