Computational and Structural Biotechnology Journal ( IF 4.4 ) Pub Date : 2019-12-27 , DOI: 10.1016/j.csbj.2019.12.008 Suyu Mei 1 , Kun Zhang 2
|
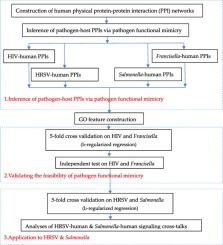
Pathogen-host protein interactions are fundamental for pathogens to manipulate host signaling pathways and subvert host immune defense. For most pathogens, very few or no experimental studies have been conducted to investigate their signaling cross-talks with host. In this study, we propose a computational framework to validate the biological assumption that human protein–protein interaction (PPI) networks alone are sufficient to infer pathogen-host PPIs via pathogen functional mimicry. Pathogen functional mimicry assumes that a pathogen functionally mimics and substitutes host counterpart proteins in order for the pathogen to get involved in or hijack the host cellular processes. Through pathogen functional mimicry defined via gene ontology (GO) semantic similarity, we first use the known human PPIs as templates to infer pathogen-host PPIs, and the PPIs are further used as training data to build an l2-regularized logistic regression model for novel pathogen-host PPI prediction. Independent tests on the experimental data from human immunodeficiency virus and Francisella tularensis validate the effectiveness of the proposed pathogen functional mimicry technique. Performance comparisons also show that the proposed technique y excels the existing pathogen sequence mimicry approaches and transfer learning methods. The proposed framework provides a new avenue to study the experimentally less-studied pathogens in the worst scenarios that very few or no experimental pathogen-host PPIs are available. As two case studies, we apply the proposed framework to Salmonella typhimurium and Human respiratory syncytial virus to reconstruct the pathogen-host PPI networks and further investigate the interference of these two pathogens with human immune signaling and transcription regulatory system.
中文翻译:

通过病原体模拟和人类蛋白质-蛋白质相互作用网络在计算机上解开病原体-宿主信号传导的串扰。
病原体-宿主蛋白相互作用是病原体操纵宿主信号通路和破坏宿主免疫防御的基础。对于大多数病原体,很少或没有进行实验研究来研究它们与宿主的信号交叉对话。在这项研究中,我们提出了一个计算框架来验证生物学假设,即人类蛋白质 - 蛋白质相互作用(PPI)网络足以通过病原体功能模拟推断病原体 - 宿主 PPI。病原体功能模拟假设病原体在功能上模拟和替代宿主对应蛋白,以使病原体参与或劫持宿主细胞过程。通过通过基因本体(GO)语义相似性定义的病原体功能模拟,我们首先使用已知的人类 PPI 作为模板来推断病原体-宿主 PPI,用于新病原体-宿主 PPI 预测的2正则化逻辑回归模型。对人类免疫缺陷病毒和土拉弗朗西斯菌实验数据的独立测试验证了所提出的病原体功能模拟技术的有效性。性能比较还表明,所提出的技术 y 优于现有的病原体序列模拟方法和迁移学习方法。所提出的框架提供了一条新途径,可以在极少或没有实验性病原体宿主 PPI 可用的最坏情况下研究实验性研究较少的病原体。作为两个案例研究,我们将提出的框架应用于鼠伤寒沙门氏菌和人类呼吸道合胞病毒重建病原体-宿主PPI网络,进一步研究这两种病原体对人体免疫信号和转录调控系统的干扰。









































 京公网安备 11010802027423号
京公网安备 11010802027423号