Computational and Structural Biotechnology Journal ( IF 4.4 ) Pub Date : 2019-12-27 , DOI: 10.1016/j.csbj.2019.12.009 Chia-Yin Chiang , Yung-Hao Ching , Ting-Yan Chang , Liang-Shuan Hu , Yee Siang Yong , Pei Ying Keak , Ivana Mustika , Ming-Der Lin , Ben-Yang Liao
|
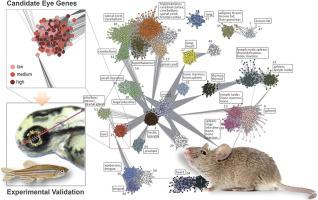
In the last few decades, reverse genetic and high throughput approaches have been frequently applied to the mouse (Mus musculus) to understand how genes function in tissues/organs and during development in a mammalian system. Despite these efforts, the associated phenotypes for the majority of mouse genes remained to be fully characterized. Here, we performed an integrated transcriptome-phenome analysis by identifying coexpressed gene modules based on tissue transcriptomes profiled with each of various platforms and functionally interpreting these modules using the mouse phenotypic data. Consequently, >15,000 mouse genes were linked with at least one of the 47 tissue functions that were examined. Specifically, our approach predicted >50 genes previously unknown to be involved in mice (Mus musculus) visual functions. Fifteen genes were selected for further analysis based on their potential biomedical relevance and compatibility with further experimental validation. Gene-specific morpholinos were introduced into zebrafish (Danio rerio) to target their corresponding orthologs. Quantitative assessments of phenotypes of developing eyes confirmed predicted eye-related functions of 13 out of the 15 genes examined. These novel eye genes include: Adal, Ankrd33, Car14, Ccdc126, Dhx32, Dkk3, Fam169a, Grifin, Kcnj14, Lrit2, Ppef2, Ppm1n, and Wdr17. The results highlighted the potential for this phenome-based approach to assist the experimental design of mutating and phenotyping mouse genes that aims to fully reveal the functional landscape of mammalian genomes.
中文翻译:

通过对小鼠转录组和表型的综合分析系统地发现了新的眼睛基因。
在过去的几十年中,反向遗传学和高通量方法已经常应用于小鼠(小家鼠),以了解基因在组织/器官中的功能以及哺乳动物系统发育过程中的功能。尽管做出了这些努力,但大多数小鼠基因的相关表型仍有待充分表征。在这里,我们通过基于与各种平台中的每一个分析的组织转录组确定共表达的基因模块,并使用小鼠表型数据功能性地解释了这些模块,从而进行了整合的转录组-现象分析。因此,> 15,000个小鼠基因与所检查的47种组织功能中的至少一种相关。具体而言,我们的方法预测了超过50个以前未知的基因与小鼠(Mus musculus)有关。)视觉功能。根据其潜在的生物医学相关性和与进一步实验验证的兼容性,选择了15个基因进行进一步分析。将基因特异的吗啉代引入斑马鱼(Danio rerio)中,以靶向其相应的直系同源物。对发育中的眼睛表型的定量评估证实了所检查的15个基因中有13个具有预测的与眼睛相关的功能。这些新颖的眼睛基因包括:Adal,Ankrd33,Car14,Ccdc126,Dhx32,Dkk3,Fam169a,Grifin,Kcnj14,Lrit2,Ppef2,Ppm1n和Wdr17。结果强调了这种基于表型的方法可能有助于突变和表型小鼠基因实验设计的目的,该实验旨在充分揭示哺乳动物基因组的功能格局。











































 京公网安备 11010802027423号
京公网安备 11010802027423号