当前位置:
X-MOL 学术
›
Mol. Omics
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Mass spectrometric analysis of core fucosylation and sequence variation in a human-camelid monoclonal antibody.
Molecular Omics ( IF 3.0 ) Pub Date : 2020-03-04 , DOI: 10.1039/c9mo00168a Lynda J Donald 1 , Maureen Spearman , Neha Mishra , Emy Komatsu , Michael Butler , Hélène Perreault
Molecular Omics ( IF 3.0 ) Pub Date : 2020-03-04 , DOI: 10.1039/c9mo00168a Lynda J Donald 1 , Maureen Spearman , Neha Mishra , Emy Komatsu , Michael Butler , Hélène Perreault
Affiliation
|
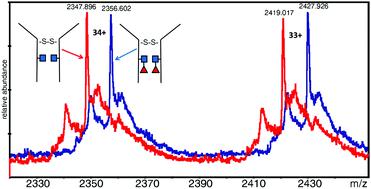
Electrospray mass spectrometry (ESI-MS) was used to measure the masses of an intact dimeric monoclonal antibody (Mab) and assess the fucosylation level. The Mab under study was EG2-hFc, a chimeric human–camelid antibody of about 80 kDa (A. Bell et al., Cancer Lett., 2010, 289(1), 81–90). It was obtained from cell culture with and without a fucosylation inhibitor, and treated with EndoS which cleaves between the two core N-acetyl glucosamine (GlcNAc) residues. It is the first time that this combined approach with a unique mass spectrometer was used to measure 146 Da differences as part of a large intact dimeric antibody. Results showed that in the dimer, both heavy chains were fucosylated on the core GlcNAc of the Fc Asn site equivalent to Asn297. In the presence of the fucosylation inhibitor, fucosylation was lost on both subunits. Following reduction, monomers were analyzed and the masses obtained corroborated the dimer results. Dimeric EG2-hFc Mab treated with PNGase F, to deglycosylate the protein, was also measured by MS for mass comparison. In spite of the success of fucosylation level measurements, the experimental masses of deglycosylated dimers and GlcNAc–Fuc bearing dimers did not correspond to masses of our sequence of reference (A. Bell et al., Cancer Lett., 2010, 289(1), 81–90; http://www.uniprot.org; http://www.expasy.org), which prompted experiments to determine the protein backbone sequence. Digest mixtures from trypsin, GluC, as well as trypsin + GluC proteolysis were analyzed by matrix-assisted laser desorption/ionization (MALDI) MS and MS/MS. A few variations were found relative to the reference sequence, which are discussed in detail herein. These measurements allowed us to build a new “experimental” sequence for the EG2-hFc samples investigated in this work, although there are still ambiguities to be resolved in this new sequence. MALDI-MS/MS also confirmed the fucosylation pattern in the Fc tryptic peptide EEQYNSTYR.
中文翻译:

质谱分析人骆驼科单克隆抗体中的岩藻糖基化和序列变异。
电喷雾质谱法(ESI-MS)用于测量完整的二聚体单克隆抗体(Mab)的质量并评估岩藻糖基化水平。被研究的Mab是EG2-hFc,一种约80 kDa的嵌合人-骆驼状抗体(A. Bell等人,Cancer Lett。,2010,289(1),81-90)。它是从有或没有岩藻糖基化抑制剂的细胞培养物中获得的,并用在两个核心N之间裂解的EndoS处理-乙酰基葡糖胺(GlcNAc)残基。这是首次将这种结合独特质谱仪的方法用于测量146 Da差异,作为完整的二聚抗体的一部分。结果显示,在二聚体中,两条重链都在与Asn297相同的Fc Asn位点的核心GlcNAc上被岩藻糖基化。在岩藻糖基化抑制剂的存在下,岩藻糖基化在两个亚基上均丢失。还原后,分析单体,得到的质量证实了二聚体结果。还通过MS测量了用PNGase F处理以使蛋白质去糖基化的二聚体EG2-hFc Mab,以进行质量比较。尽管岩藻糖基化水平测量成功,但去糖基化二聚体和带有GlcNAc-Fuc的二聚体的实验质量与我们参考序列的质量不符(A. Bell等。,巨蟹座Lett。,2010,289(1),81–90; http://www.uniprot.org; http://www.expasy.org),这促使实验确定蛋白质骨架序列。通过基质辅助激光解吸/电离(MALDI)MS和MS / MS分析了来自胰蛋白酶,GluC以及胰蛋白酶+ GluC蛋白水解的消化混合物。相对于参考序列,发现了一些变体,在此详细讨论。这些测量结果使我们能够为这项工作中研究的EG2-hFc样品建立一个新的“实验”序列,尽管在这个新序列中仍然有待解决的歧义。MALDI-MS / MS还证实了Fc胰蛋白酶肽EEQYNSTYR中的岩藻糖基化模式。
更新日期:2020-03-04
中文翻译:

质谱分析人骆驼科单克隆抗体中的岩藻糖基化和序列变异。
电喷雾质谱法(ESI-MS)用于测量完整的二聚体单克隆抗体(Mab)的质量并评估岩藻糖基化水平。被研究的Mab是EG2-hFc,一种约80 kDa的嵌合人-骆驼状抗体(A. Bell等人,Cancer Lett。,2010,289(1),81-90)。它是从有或没有岩藻糖基化抑制剂的细胞培养物中获得的,并用在两个核心N之间裂解的EndoS处理-乙酰基葡糖胺(GlcNAc)残基。这是首次将这种结合独特质谱仪的方法用于测量146 Da差异,作为完整的二聚抗体的一部分。结果显示,在二聚体中,两条重链都在与Asn297相同的Fc Asn位点的核心GlcNAc上被岩藻糖基化。在岩藻糖基化抑制剂的存在下,岩藻糖基化在两个亚基上均丢失。还原后,分析单体,得到的质量证实了二聚体结果。还通过MS测量了用PNGase F处理以使蛋白质去糖基化的二聚体EG2-hFc Mab,以进行质量比较。尽管岩藻糖基化水平测量成功,但去糖基化二聚体和带有GlcNAc-Fuc的二聚体的实验质量与我们参考序列的质量不符(A. Bell等。,巨蟹座Lett。,2010,289(1),81–90; http://www.uniprot.org; http://www.expasy.org),这促使实验确定蛋白质骨架序列。通过基质辅助激光解吸/电离(MALDI)MS和MS / MS分析了来自胰蛋白酶,GluC以及胰蛋白酶+ GluC蛋白水解的消化混合物。相对于参考序列,发现了一些变体,在此详细讨论。这些测量结果使我们能够为这项工作中研究的EG2-hFc样品建立一个新的“实验”序列,尽管在这个新序列中仍然有待解决的歧义。MALDI-MS / MS还证实了Fc胰蛋白酶肽EEQYNSTYR中的岩藻糖基化模式。











































 京公网安备 11010802027423号
京公网安备 11010802027423号