当前位置:
X-MOL 学术
›
Mol. Phylogenet. Evol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Phylogenetic relationships and chloroplast capture in the Amelanchier-Malacomeles-Peraphyllum clade (Maleae, Rosaceae): Evidence from chloroplast genome and nuclear ribosomal DNA data using genome skimming.
Molecular Phylogenetics and Evolution ( IF 4.1 ) Pub Date : 2020-03-02 , DOI: 10.1016/j.ympev.2020.106784 Bin-Bin Liu 1 , Christopher S Campbell 2 , De-Yuan Hong 3 , Jun Wen 4
Molecular Phylogenetics and Evolution ( IF 4.1 ) Pub Date : 2020-03-02 , DOI: 10.1016/j.ympev.2020.106784 Bin-Bin Liu 1 , Christopher S Campbell 2 , De-Yuan Hong 3 , Jun Wen 4
Affiliation
|
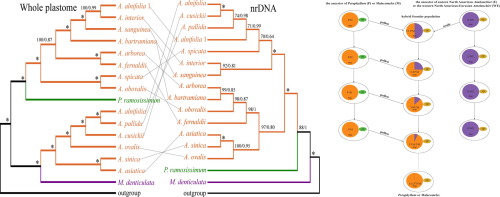
The Amelanchier-Malacomeles-Peraphyllum (AMP) clade consists of ca. 26 species distributed in North and Central America, Europe, Asia, and northwestern Africa. While molecular and morphological data strongly support this clade, relationships of its genera are uncertain. Support for the monophyly of Amelanchier and for the phylogenetic positions of Malacomeles and Peraphyllum has varied between studies. Our goals were to reconstruct a robust phylogeny of the AMP clade in the framework of Maleae and clarify the phylogenetic placements of Malacomeles and Peraphyllum. This study employs sequences of the whole plastome and nuclear ribosomal DNA (nrDNA) repeats assembled using genome skimming with 131 samples representing 115 species in 31 genera of Rosaceae, especially Maleae. Maximum likelihood (ML) and Bayesian analysis (BI) of whole plastome datasets strongly supported Amelanchier as not monophyletic, with Peraphyllum sister to eastern North American Amelanchier and Malacomeles sister to the western North American-Eurasian Amelanchier. In contrast, nrDNA recovered the monophyly of Amelanchier, with Peraphyllum sister to Amelanchier and Malacomeles sister to the Amelanchier-Peraphyllum clade. The strong topological conflicts between plastome and nrDNA phylogenies of Peraphyllum and of Malacomeles are best explained by ancient chloroplast capture that occurred in SW North America.
中文翻译:

系统发育关系和叶绿体捕获在Amelanchier-Malacomeles-Peraphyllum进化枝(马累,蔷薇科)中:来自叶绿体基因组和核糖核体DNA数据的证据,使用基因组略读。
Amelanchier-Malacomeles-Peraphyllum(AMP)进化枝由大约 26个物种分布在北美和中美洲,欧洲,亚洲和西北非洲。尽管分子和形态学数据强烈支持这一进化枝,但其属的关系尚不确定。在研究之间,对Amelanchier的单方面研究以及对Malacomeles和Peraphyllum的系统发生地位的支持有所不同。我们的目标是在马来亚框架内重建AMP进化枝的稳固系统发育,并阐明马拉科梅勒和Peraphyllum的系统发生位置。这项研究使用了整个质体组和核糖体DNA(nrDNA)重复序列,这些重复序列是通过基因组略读组装而成的,共包含131种样品,这些样品代表了蔷薇科(尤其是马来亚科)的31个属。整个质体组数据集的最大似然(ML)和贝叶斯分析(BI)强烈支持Amelanchier不具有单元性,Peraphyllum姐姐是北美东部Amelanchier的姊妹,而Malacomeles姐姐则是北美西部-欧亚Amelanchier的姊妹。相比之下,nrDNA恢复了Amelanchier的单亲性,Peraphyllum姐姐是Amelanchier的姐妹,而Malacomeles姐姐则是Amelanchier-Peraphyllum进化枝的姐妹。百草枯和马拉科麦勒的质体组和nrDNA系统发育之间强烈的拓扑冲突可以用发生在北美西南部的古代叶绿体捕获来很好地解释。nrDNA回收了Amelanchier的单亲生物,Peraphyllum姐妹为Amelanchier,Malacomeles姐妹为Amelanchier-Peraphyllum进化枝。百草枯和马拉科麦勒的质体组和nrDNA系统发育之间强烈的拓扑冲突可以用发生在北美西南部的古代叶绿体捕获来很好地解释。nrDNA回收了Amelanchier的单亲生物,Peraphyllum姐妹为Amelanchier,Malacomeles姐妹为Amelanchier-Peraphyllum进化枝。百草枯和马拉科麦勒的质体组和nrDNA系统发育之间强烈的拓扑冲突可以用发生在北美西南部的古代叶绿体捕获来很好地解释。
更新日期:2020-03-03
中文翻译:

系统发育关系和叶绿体捕获在Amelanchier-Malacomeles-Peraphyllum进化枝(马累,蔷薇科)中:来自叶绿体基因组和核糖核体DNA数据的证据,使用基因组略读。
Amelanchier-Malacomeles-Peraphyllum(AMP)进化枝由大约 26个物种分布在北美和中美洲,欧洲,亚洲和西北非洲。尽管分子和形态学数据强烈支持这一进化枝,但其属的关系尚不确定。在研究之间,对Amelanchier的单方面研究以及对Malacomeles和Peraphyllum的系统发生地位的支持有所不同。我们的目标是在马来亚框架内重建AMP进化枝的稳固系统发育,并阐明马拉科梅勒和Peraphyllum的系统发生位置。这项研究使用了整个质体组和核糖体DNA(nrDNA)重复序列,这些重复序列是通过基因组略读组装而成的,共包含131种样品,这些样品代表了蔷薇科(尤其是马来亚科)的31个属。整个质体组数据集的最大似然(ML)和贝叶斯分析(BI)强烈支持Amelanchier不具有单元性,Peraphyllum姐姐是北美东部Amelanchier的姊妹,而Malacomeles姐姐则是北美西部-欧亚Amelanchier的姊妹。相比之下,nrDNA恢复了Amelanchier的单亲性,Peraphyllum姐姐是Amelanchier的姐妹,而Malacomeles姐姐则是Amelanchier-Peraphyllum进化枝的姐妹。百草枯和马拉科麦勒的质体组和nrDNA系统发育之间强烈的拓扑冲突可以用发生在北美西南部的古代叶绿体捕获来很好地解释。nrDNA回收了Amelanchier的单亲生物,Peraphyllum姐妹为Amelanchier,Malacomeles姐妹为Amelanchier-Peraphyllum进化枝。百草枯和马拉科麦勒的质体组和nrDNA系统发育之间强烈的拓扑冲突可以用发生在北美西南部的古代叶绿体捕获来很好地解释。nrDNA回收了Amelanchier的单亲生物,Peraphyllum姐妹为Amelanchier,Malacomeles姐妹为Amelanchier-Peraphyllum进化枝。百草枯和马拉科麦勒的质体组和nrDNA系统发育之间强烈的拓扑冲突可以用发生在北美西南部的古代叶绿体捕获来很好地解释。



























 京公网安备 11010802027423号
京公网安备 11010802027423号