当前位置:
X-MOL 学术
›
J. Mol. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Unraveling Allostery in a Knotted Minimal Methyltransferase by NMR Spectroscopy.
Journal of Molecular Biology ( IF 4.7 ) Pub Date : 2020-03-02 , DOI: 10.1016/j.jmb.2020.02.029 Dominique T Capraro 1 , David J Burban 1 , Patricia A Jennings 1
Journal of Molecular Biology ( IF 4.7 ) Pub Date : 2020-03-02 , DOI: 10.1016/j.jmb.2020.02.029 Dominique T Capraro 1 , David J Burban 1 , Patricia A Jennings 1
Affiliation
|
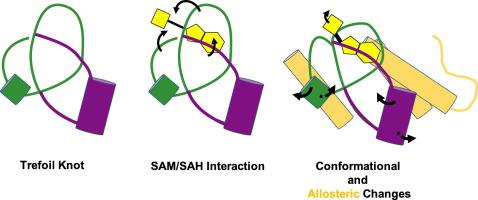
The methyltransferases that belong to the SpoU-TrmD family contain trefoil knots in their backbone fold. Recent structural dynamic and binding analyses of both free and bound homologs indicate that the knot within the polypeptide backbone plays a significant role in the biological activity of the molecule. The knot loops form the S-adenosyl-methionine (SAM)-binding pocket as well as participate in SAM binding and catalysis. Knots contain both at once a stable core as well as moving parts that modulate long-range motions. Here, we sought to understand allosteric effects modulated by the knotted topology. Uncovering the residues that contribute to these changes and the functional aspects of these protein motions are essential to understanding the interplay between the knot, activation of the methyltransferase, and the implications in RNA interactions. The question we sought to address is as follows: How does the knot, which constricts the backbone as well as forms the SAM-binding pocket with its three distinctive loops, affect the binding mechanism? Using a minimally tied trefoil protein as the framework for understanding the structure-function roles, we offer an unprecedented view of the conformational mechanics of the knot and its relationship to the activation of the ligand molecule. Focusing on the biophysical characterization of the knot region by NMR spectroscopy, we identify the SAM-binding region and observe changes in the dynamics of the loops that form the knot. Importantly, we also observe long-range allosteric changes in flanking helices consistent with winding/unwinding in helical propensity as the knot tightens to secure the SAM cofactor.
中文翻译:

通过核磁共振波谱揭示打结的最小甲基转移酶的变构。
属于 SpoU-TrmD 家族的甲基转移酶在其主链折叠中含有三叶结。最近对游离和结合同系物的结构动态和结合分析表明,多肽主链内的结在分子的生物活性中起着重要作用。结环形成 S-腺苷甲硫氨酸 (SAM) 结合口袋并参与 SAM 结合和催化。结既包含稳定的核心,也包含调节长距离运动的运动部件。在这里,我们试图了解打结拓扑调节的变构效应。揭示导致这些变化的残基以及这些蛋白质运动的功能方面对于理解结之间的相互作用、甲基转移酶的激活以及 RNA 相互作用的影响至关重要。我们试图解决的问题如下:结收缩主链并形成具有三个独特环的 SAM 结合袋,它如何影响结合机制?使用最小化三叶蛋白作为理解结构功能作用的框架,我们提供了结的构象力学及其与配体分子激活的关系的前所未有的观点。我们通过核磁共振波谱法重点关注结区域的生物物理表征,识别 SAM 结合区域并观察形成结的环的动力学变化。重要的是,我们还观察到侧翼螺旋的长程变构变化与当结拉紧以固定 SAM 辅因子时螺旋倾向的缠绕/解开一致。
更新日期:2020-03-02
中文翻译:

通过核磁共振波谱揭示打结的最小甲基转移酶的变构。
属于 SpoU-TrmD 家族的甲基转移酶在其主链折叠中含有三叶结。最近对游离和结合同系物的结构动态和结合分析表明,多肽主链内的结在分子的生物活性中起着重要作用。结环形成 S-腺苷甲硫氨酸 (SAM) 结合口袋并参与 SAM 结合和催化。结既包含稳定的核心,也包含调节长距离运动的运动部件。在这里,我们试图了解打结拓扑调节的变构效应。揭示导致这些变化的残基以及这些蛋白质运动的功能方面对于理解结之间的相互作用、甲基转移酶的激活以及 RNA 相互作用的影响至关重要。我们试图解决的问题如下:结收缩主链并形成具有三个独特环的 SAM 结合袋,它如何影响结合机制?使用最小化三叶蛋白作为理解结构功能作用的框架,我们提供了结的构象力学及其与配体分子激活的关系的前所未有的观点。我们通过核磁共振波谱法重点关注结区域的生物物理表征,识别 SAM 结合区域并观察形成结的环的动力学变化。重要的是,我们还观察到侧翼螺旋的长程变构变化与当结拉紧以固定 SAM 辅因子时螺旋倾向的缠绕/解开一致。











































 京公网安备 11010802027423号
京公网安备 11010802027423号