Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics ( IF 2.5 ) Pub Date : 2020-03-02 , DOI: 10.1016/j.bbapap.2020.140406 Chaolu Meng 1 , Jun Zhang 2 , Xiucai Ye 3 , Fei Guo 4 , Quan Zou 5
|
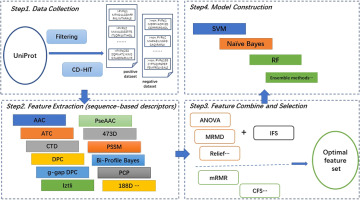
Phage virion protein (PVP) identification plays key role in elucidating relationships between phages and hosts. Moreover, PVP identification can facilitate the design of related biochemical entities. Recently, several machine learning approaches have emerged for this purpose and have shown their potential capacities. In this study, the proposed PVP identifiers are systemically reviewed, and the related algorithms and tools are comprehensively analyzed. We summarized the common framework of these PVP identifiers and constructed our own novel identifiers based upon the framework. Furthermore, we focus on a performance comparison of all PVP identifiers by using a training dataset and an independent dataset. Highlighting the pros and cons of these identifiers demonstrates that g-gap DPC (dipeptide composition) features are capable of representing characteristics of PVPs. Moreover, SVM (support vector machine) is proven to be the more effective classifier to distinguish PVPs and non-PVPs.
中文翻译:

综述和比较基于机器学习的噬菌体病毒粒子蛋白质鉴定方法。
噬菌体病毒粒子蛋白(PVP)的鉴定在阐明噬菌体与宿主之间的关系中起着关键作用。而且,PVP鉴定可以促进相关生化实体的设计。最近,为此目的出现了几种机器学习方法,并显示了它们的潜在能力。在这项研究中,系统地审查了提出的PVP标识符,并全面分析了相关的算法和工具。我们总结了这些PVP标识符的通用框架,并基于该框架构造了我们自己的新颖标识符。此外,我们专注于通过使用训练数据集和独立数据集比较所有PVP标识符的性能。强调这些标识符的优缺点表明,g-gap DPC(二肽组成)功能能够代表PVP的特征。此外,事实证明,SVM(支持向量机)是区分PVP和非PVP的更有效的分类器。










































 京公网安备 11010802027423号
京公网安备 11010802027423号