Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Parentage and relatedness reconstruction in Pinus sylvestris using genotyping-by-sequencing
Heredity ( IF 3.1 ) Pub Date : 2020-03-02 , DOI: 10.1038/s41437-020-0302-3 David Hall 1 , Wei Zhao 1, 2 , Ulfstand Wennström 3 , Bengt Andersson Gull 3 , Xiao-Ru Wang 1
Heredity ( IF 3.1 ) Pub Date : 2020-03-02 , DOI: 10.1038/s41437-020-0302-3 David Hall 1 , Wei Zhao 1, 2 , Ulfstand Wennström 3 , Bengt Andersson Gull 3 , Xiao-Ru Wang 1
Affiliation
|
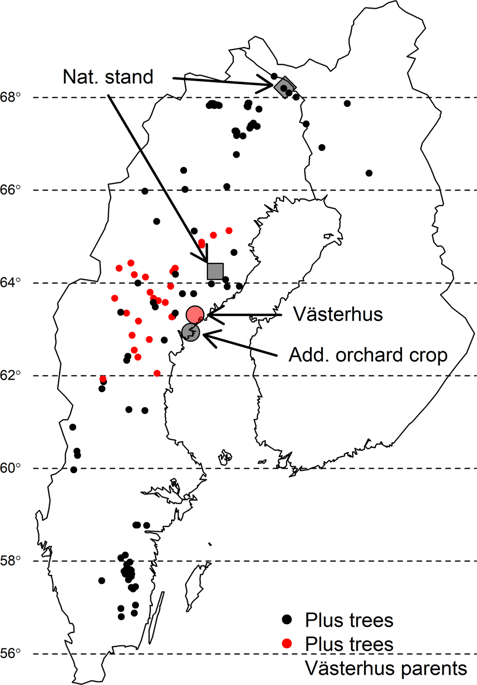
Estimating kinship is fundamental for studies of evolution, conservation, and breeding. Genotyping-by-sequencing (GBS) and other restriction based genotyping methods have become widely applied in these applications in non-model organisms. However, sequencing errors, depth, and reproducibility between library preps could potentially hinder accurate genetic inferences. In this study, we tested different sets of parameters in data filtering, different reference populations and eight estimation methods to obtain a robust procedure for relatedness estimation in Scots pine (Pinus sylvestris L.). We used a seed orchard as our study system, where candidate parents are known and pedigree reconstruction can be compared with theoretical expectations. We found that relatedness estimates were lower than expected for all categories of kinship estimated if the proportion of shared SNPs was low. However, estimates reached expected values if loci showing an excess of heterozygotes were removed and genotyping error rates were considered. The genetic variance-covariance matrix (G-matrix) estimation, however, performed poorly in kinship estimation. The reduced relatedness estimates are likely due to false heterozygosity calls. We analyzed the mating structure in the seed orchard and identified a selfing rate of 3% (including crosses between clone mates) and external pollen contamination of 33.6%. Little genetic structure was observed in the sampled Scots pine natural populations, and the degree of inbreeding in the orchard seed crop is comparable to natural stands. We illustrate that under our optimized data processing procedure, relatedness, and genetic composition, including level of pollen contamination within a seed orchard crop, can be established consistently by different estimators.
中文翻译:

使用基因分型测序法重建樟子松的亲缘关系和亲缘关系
估计亲属关系是进化、保护和育种研究的基础。基于测序的基因分型 (GBS) 和其他基于限制的基因分型方法已广泛应用于非模式生物的这些应用中。然而,文库制备之间的测序错误、深度和可重复性可能会阻碍准确的遗传推断。在这项研究中,我们测试了数据过滤中的不同参数集、不同的参考种群和八种估计方法,以获得用于苏格兰松 (Pinus sylvestris L.) 相关性估计的稳健程序。我们使用种子园作为我们的研究系统,其中候选父母是已知的,并且可以将谱系重建与理论预期进行比较。我们发现,如果共享 SNP 的比例较低,则所有亲属关系类别的相关性估计都低于预期。然而,如果去除显示过量杂合子的基因座并考虑基因分型错误率,则估计值达到预期值。然而,遗传方差-协方差矩阵(G-矩阵)估计在亲属关系估计中表现不佳。减少的相关性估计可能是由于错误的杂合性调用。我们分析了种子园的交配结构并确定了 3% 的自交率(包括克隆配偶之间的杂交)和 33.6% 的外部花粉污染。在抽样的苏格兰松自然种群中几乎没有观察到遗传结构,果园种子作物的近交程度与自然林相当。
更新日期:2020-03-02
中文翻译:

使用基因分型测序法重建樟子松的亲缘关系和亲缘关系
估计亲属关系是进化、保护和育种研究的基础。基于测序的基因分型 (GBS) 和其他基于限制的基因分型方法已广泛应用于非模式生物的这些应用中。然而,文库制备之间的测序错误、深度和可重复性可能会阻碍准确的遗传推断。在这项研究中,我们测试了数据过滤中的不同参数集、不同的参考种群和八种估计方法,以获得用于苏格兰松 (Pinus sylvestris L.) 相关性估计的稳健程序。我们使用种子园作为我们的研究系统,其中候选父母是已知的,并且可以将谱系重建与理论预期进行比较。我们发现,如果共享 SNP 的比例较低,则所有亲属关系类别的相关性估计都低于预期。然而,如果去除显示过量杂合子的基因座并考虑基因分型错误率,则估计值达到预期值。然而,遗传方差-协方差矩阵(G-矩阵)估计在亲属关系估计中表现不佳。减少的相关性估计可能是由于错误的杂合性调用。我们分析了种子园的交配结构并确定了 3% 的自交率(包括克隆配偶之间的杂交)和 33.6% 的外部花粉污染。在抽样的苏格兰松自然种群中几乎没有观察到遗传结构,果园种子作物的近交程度与自然林相当。









































 京公网安备 11010802027423号
京公网安备 11010802027423号