当前位置:
X-MOL 学术
›
ACS Chem. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
ACS Chemical Biology ( IF 3.5 ) Pub Date : 2020-03-02 , DOI: 10.1021/acschembio.9b00894 Rajiv Kumar Bedi 1 , Danzhi Huang 1 , Lars Wiedmer 1 , Yaozong Li 1 , Aymeric Dolbois 1 , Justyna Aleksandra Wojdyla 2 , May Elizabeth Sharpe 2 , Amedeo Caflisch 1 , Pawel Sledz 1
ACS Chemical Biology ( IF 3.5 ) Pub Date : 2020-03-02 , DOI: 10.1021/acschembio.9b00894 Rajiv Kumar Bedi 1 , Danzhi Huang 1 , Lars Wiedmer 1 , Yaozong Li 1 , Aymeric Dolbois 1 , Justyna Aleksandra Wojdyla 2 , May Elizabeth Sharpe 2 , Amedeo Caflisch 1 , Pawel Sledz 1
Affiliation
|
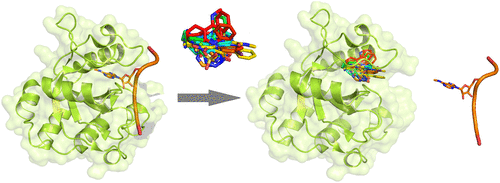
We report a crystallographic analysis of small-molecule ligands of the human YTHDC1 domain that recognizes N6-methylated adenine (m6A) in RNA. The 30 binders are fragments (molecular weight < 300 g mol-1) that represent 10 different chemotypes identified by virtual screening. Despite the structural disorder of the binding site loop (residues 429-439), most of the 30 fragments emulate the two main interactions of the -NHCH3 group of m6A. These interactions are the hydrogen bond to the backbone carbonyl of Ser378 and the van der Waals contacts with the tryptophan cage. Different chemical groups are involved in the conserved binding motifs. Some of the fragments show favorable ligand efficiency for YTHDC1 and selectivity against other m6A reader domains. The structural information is useful for the design of modulators of m6A recognition by YTHDC1.
中文翻译:

选择性破坏m6A依赖性蛋白RNA与片段的相互作用。
我们报告人YTHDC1域的小分子配体的晶体学分析,该小分子配体识别RNA中的N6-甲基化腺嘌呤(m6A)。30种结合剂是片段(分子量<300 g mol-1),代表通过虚拟筛选鉴定出的10种不同的化学型。尽管存在结合位点环的结构异常(残基429-439),但30个片段中的大多数模拟了m6A的-NHCH3基团的两个主要相互作用。这些相互作用是Ser378的骨架羰基的氢键,范德华斯与色氨酸笼子接触。保守的结合基序涉及不同的化学基团。一些片段显示出对YTHDC1有利的配体效率和对其他m6A读者域的选择性。结构信息对于YTHDC1识别m6A的调节剂的设计很有用。
更新日期:2020-03-03
中文翻译:

选择性破坏m6A依赖性蛋白RNA与片段的相互作用。
我们报告人YTHDC1域的小分子配体的晶体学分析,该小分子配体识别RNA中的N6-甲基化腺嘌呤(m6A)。30种结合剂是片段(分子量<300 g mol-1),代表通过虚拟筛选鉴定出的10种不同的化学型。尽管存在结合位点环的结构异常(残基429-439),但30个片段中的大多数模拟了m6A的-NHCH3基团的两个主要相互作用。这些相互作用是Ser378的骨架羰基的氢键,范德华斯与色氨酸笼子接触。保守的结合基序涉及不同的化学基团。一些片段显示出对YTHDC1有利的配体效率和对其他m6A读者域的选择性。结构信息对于YTHDC1识别m6A的调节剂的设计很有用。











































 京公网安备 11010802027423号
京公网安备 11010802027423号