Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Crystal Structure of the Mannose-6-Phosphate Uncovering Enzyme.
Structure ( IF 4.4 ) Pub Date : 2020-02-27 , DOI: 10.1016/j.str.2020.02.001 Alexei Gorelik 1 , Katalin Illes 1 , Bhushan Nagar 1
Structure ( IF 4.4 ) Pub Date : 2020-02-27 , DOI: 10.1016/j.str.2020.02.001 Alexei Gorelik 1 , Katalin Illes 1 , Bhushan Nagar 1
Affiliation
|
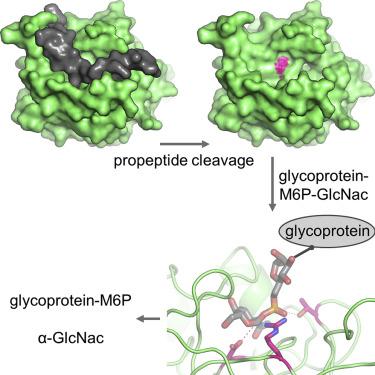
Most lysosomal hydrolytic enzymes reach their destination via the mannose-6-phosphate (M6P) pathway. The enzyme N-acetylglucosamine-1-phosphodiester α-N-acetylglucosaminidase (NAGPA, or "uncovering enzyme") catalyzes the second step in the M6P tag formation, namely the removal of the masking N-acetylglucosamine (GlcNAc) portion. Defects in this protein are associated with non-syndromic stuttering. To gain a better understanding of the function and regulation of this enzyme, we determined its crystal structure. The propeptide binds in a groove on the globular catalytic domain, blocking active site access. High-affinity substrate binding is enabled by a conformational switch in an active site loop. The protein recognizes the GlcNAc and phosphate portions of its substrate, but not the mannose moiety of the glycan. Based on enzymatic and 1H-NMR analysis, a catalytic mechanism is proposed. Crystallographic and solution scattering analyses suggest that the C-terminal domain forms a long flexible stem that extends the enzyme away from the Golgi membrane.
中文翻译:

甘露糖6磷酸揭解酶的晶体结构。
大多数溶酶体水解酶通过6-磷酸甘露糖(M6P)途径到达目的地。N-乙酰氨基葡萄糖-1-磷酸二酯酶α-N-乙酰氨基葡萄糖苷酶(NAGPA或“未覆盖酶”)催化M6P标签形成的第二步,即去除掩蔽的N-乙酰氨基葡萄糖(GlcNAc)部分。该蛋白的缺陷与非综合征性口吃有关。为了更好地了解该酶的功能和调控,我们确定了其晶体结构。前肽结合在球形催化结构域上的凹槽中,阻止了活性位点的进入。高亲和力的底物结合是通过活性位点环中的构象开关实现的。该蛋白质识别其底物的GlcNAc和磷酸部分,但不识别聚糖的甘露糖部分。基于酶和1 H-NMR分析,提出了催化机理。晶体学和溶液散射分析表明,C-末端结构域形成长的柔性茎,该茎使酶远离高尔基体膜延伸。
更新日期:2020-02-27
中文翻译:

甘露糖6磷酸揭解酶的晶体结构。
大多数溶酶体水解酶通过6-磷酸甘露糖(M6P)途径到达目的地。N-乙酰氨基葡萄糖-1-磷酸二酯酶α-N-乙酰氨基葡萄糖苷酶(NAGPA或“未覆盖酶”)催化M6P标签形成的第二步,即去除掩蔽的N-乙酰氨基葡萄糖(GlcNAc)部分。该蛋白的缺陷与非综合征性口吃有关。为了更好地了解该酶的功能和调控,我们确定了其晶体结构。前肽结合在球形催化结构域上的凹槽中,阻止了活性位点的进入。高亲和力的底物结合是通过活性位点环中的构象开关实现的。该蛋白质识别其底物的GlcNAc和磷酸部分,但不识别聚糖的甘露糖部分。基于酶和1 H-NMR分析,提出了催化机理。晶体学和溶液散射分析表明,C-末端结构域形成长的柔性茎,该茎使酶远离高尔基体膜延伸。











































 京公网安备 11010802027423号
京公网安备 11010802027423号