当前位置:
X-MOL 学术
›
J. Biomed. Inform.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
A continuous-time Markov model approach for modeling myelodysplastic syndromes progression from cross-sectional data.
Journal of Biomedical informatics ( IF 4.5 ) Pub Date : 2020-02-26 , DOI: 10.1016/j.jbi.2020.103398 G Nicora 1 , F Moretti 1 , E Sauta 1 , M Della Porta 2 , L Malcovati 3 , M Cazzola 3 , S Quaglini 1 , R Bellazzi 1
Journal of Biomedical informatics ( IF 4.5 ) Pub Date : 2020-02-26 , DOI: 10.1016/j.jbi.2020.103398 G Nicora 1 , F Moretti 1 , E Sauta 1 , M Della Porta 2 , L Malcovati 3 , M Cazzola 3 , S Quaglini 1 , R Bellazzi 1
Affiliation
|
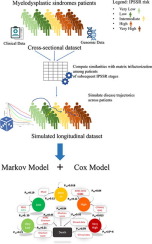
The integration of both genomics and clinical data to model disease progression is now possible, thanks to the increasing availability of molecular patients' profiles. This may lead to the definition of novel decision support tools, able to tailor therapeutic interventions on the basis of a "precise" patients' risk stratification, given their health status evolution. However, longitudinal analysis requires long-term data collection and curation, which can be time demanding, expensive and sometimes unfeasible. Here we present a clinical decision support framework that combines the simulation of disease progression from cross-sectional data with a Markov model that exploits continuous-time transition probabilities derived from Cox regression. Trajectories between patients at different disease stages are stochastically built according to a measure of patient similarity, computed with a matrix tri-factorization technique. Such trajectories are seen as realizations drawn from the stochastic process driving the transitions between the disease stages. Eventually, Markov models applied to the resulting longitudinal dataset highlight potentially relevant clinical information. We applied our method to cross-sectional genomic and clinical data from a cohort of Myelodysplastic syndromes (MDS) patients. MDS are heterogeneous clonal hematopoietic disorders whose patients are characterized by different risks of Acute Myeloid Leukemia (AML) development, defined by an international score. We computed patients' trajectories across increasing and subsequent levels of risk of developing AML, and we applied a Cox model to the simulated longitudinal dataset to assess whether genomic characteristics could be associated with a higher or lower probability of disease progression. We then used the learned parameters of such Cox model to calculate the transition probabilities of a continuous-time Markov model that describes the patients' evolution across stages. Our results are in most cases confirmed by previous studies, thus demonstrating that simulated longitudinal data represent a valuable resource to investigate disease progression of MDS patients.
中文翻译:

一种连续时间马尔可夫模型方法,用于根据横截面数据对骨髓增生异常综合症进行建模。
由于分子患者资料的可用性越来越高,现在可以将基因组学和临床数据集成到疾病进展模型中。这可能会导致定义新颖的决策支持工具,这些工具能够根据“精确的”患者风险状况(鉴于其健康状况的演变)来定制治疗干预措施。但是,纵向分析需要长期的数据收集和管理,这可能是费时,昂贵且有时不可行的。在这里,我们提出了一个临床决策支持框架,该框架将横截面数据对疾病进展的模拟与利用从Cox回归得出的连续时间转变概率的马尔可夫模型相结合。根据对患者相似性的衡量指标,随机建立了处于不同疾病阶段的患者之间的轨迹,并采用矩阵三因子技术进行了计算。这样的轨迹被视为从驱动疾病阶段之间过渡的随机过程得出的实现。最终,应用于所得纵向数据集的马尔可夫模型突出了潜在的相关临床信息。我们将我们的方法应用于来自一组骨髓增生异常综合征(MDS)患者的横断面基因组和临床数据。MDS是异质性克隆性造血系统疾病,其患者的特点是急性髓细胞性白血病(AML)发展的不同风险,这是国际评分所定义的。我们计算了发生AML的风险的增加程度及其后的患者轨迹,我们将Cox模型应用于模拟的纵向数据集,以评估基因组特征是否与疾病进展的较高或较低概率相关。然后,我们使用此类Cox模型的学习参数来计算描述患者各个阶段演变的连续时间马尔可夫模型的转移概率。在大多数情况下,我们的结果已得到先前研究的证实,因此证明了模拟的纵向数据是研究MDS患者疾病进展的宝贵资源。然后,我们使用此类Cox模型的学习参数来计算描述患者各个阶段演变的连续时间马尔可夫模型的转移概率。在大多数情况下,我们的结果已得到先前研究的证实,因此证明了模拟的纵向数据是研究MDS患者疾病进展的宝贵资源。然后,我们使用此类Cox模型的学习参数来计算描述患者各个阶段演变的连续时间马尔可夫模型的转移概率。在大多数情况下,我们的结果已得到先前研究的证实,因此证明了模拟的纵向数据是研究MDS患者疾病进展的宝贵资源。
更新日期:2020-02-26
中文翻译:

一种连续时间马尔可夫模型方法,用于根据横截面数据对骨髓增生异常综合症进行建模。
由于分子患者资料的可用性越来越高,现在可以将基因组学和临床数据集成到疾病进展模型中。这可能会导致定义新颖的决策支持工具,这些工具能够根据“精确的”患者风险状况(鉴于其健康状况的演变)来定制治疗干预措施。但是,纵向分析需要长期的数据收集和管理,这可能是费时,昂贵且有时不可行的。在这里,我们提出了一个临床决策支持框架,该框架将横截面数据对疾病进展的模拟与利用从Cox回归得出的连续时间转变概率的马尔可夫模型相结合。根据对患者相似性的衡量指标,随机建立了处于不同疾病阶段的患者之间的轨迹,并采用矩阵三因子技术进行了计算。这样的轨迹被视为从驱动疾病阶段之间过渡的随机过程得出的实现。最终,应用于所得纵向数据集的马尔可夫模型突出了潜在的相关临床信息。我们将我们的方法应用于来自一组骨髓增生异常综合征(MDS)患者的横断面基因组和临床数据。MDS是异质性克隆性造血系统疾病,其患者的特点是急性髓细胞性白血病(AML)发展的不同风险,这是国际评分所定义的。我们计算了发生AML的风险的增加程度及其后的患者轨迹,我们将Cox模型应用于模拟的纵向数据集,以评估基因组特征是否与疾病进展的较高或较低概率相关。然后,我们使用此类Cox模型的学习参数来计算描述患者各个阶段演变的连续时间马尔可夫模型的转移概率。在大多数情况下,我们的结果已得到先前研究的证实,因此证明了模拟的纵向数据是研究MDS患者疾病进展的宝贵资源。然后,我们使用此类Cox模型的学习参数来计算描述患者各个阶段演变的连续时间马尔可夫模型的转移概率。在大多数情况下,我们的结果已得到先前研究的证实,因此证明了模拟的纵向数据是研究MDS患者疾病进展的宝贵资源。然后,我们使用此类Cox模型的学习参数来计算描述患者各个阶段演变的连续时间马尔可夫模型的转移概率。在大多数情况下,我们的结果已得到先前研究的证实,因此证明了模拟的纵向数据是研究MDS患者疾病进展的宝贵资源。



























 京公网安备 11010802027423号
京公网安备 11010802027423号