Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Identifying IGH disease clones for MRD monitoring in childhood B-cell acute lymphoblastic leukemia using RNA-Seq.
Leukemia ( IF 12.8 ) Pub Date : 2020-02-25 , DOI: 10.1038/s41375-020-0774-4 Zhenhua Li 1 , Nan Jiang 1 , Evelyn Huizi Lim 1 , Winnie Hui Ni Chin 1 , Yi Lu 1 , Kean Hui Chiew 1 , Shirley Kow Yin Kham 1 , Wentao Yang 2 , Thuan Chong Quah 1, 3 , Hai Peng Lin 4 , Ah Moy Tan 5 , Hany Ariffin 6 , Jun J Yang 2 , Allen Eng-Juh Yeoh 1, 3, 7
Leukemia ( IF 12.8 ) Pub Date : 2020-02-25 , DOI: 10.1038/s41375-020-0774-4 Zhenhua Li 1 , Nan Jiang 1 , Evelyn Huizi Lim 1 , Winnie Hui Ni Chin 1 , Yi Lu 1 , Kean Hui Chiew 1 , Shirley Kow Yin Kham 1 , Wentao Yang 2 , Thuan Chong Quah 1, 3 , Hai Peng Lin 4 , Ah Moy Tan 5 , Hany Ariffin 6 , Jun J Yang 2 , Allen Eng-Juh Yeoh 1, 3, 7
Affiliation
|
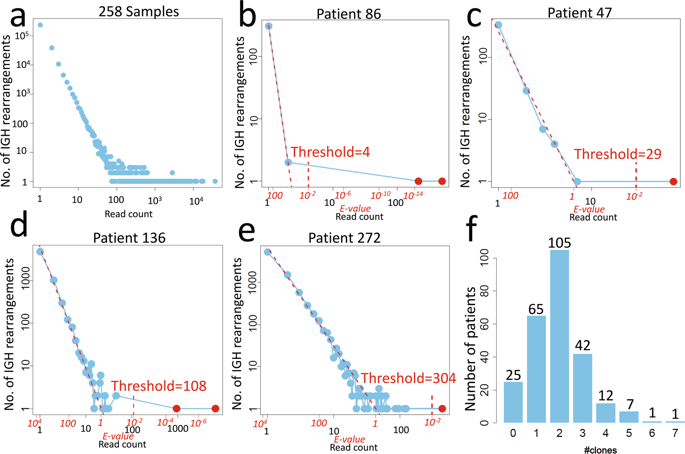
Identifying patient-specific clonal IGH/TCR junctional sequences is critical for minimal residual disease (MRD) monitoring. Conventionally these junctional sequences are identified using laborious Sanger sequencing of excised heteroduplex bands. We found that the IGH is highly expressed in our diagnostic B-cell acute lymphoblastic leukemia (B-ALL) samples using RNA-Seq. Therefore, we used RNA-Seq to identify IGH disease clone sequences in 258 childhood B-ALL samples for MRD monitoring. The amount of background IGH rearrangements uncovered by RNA-Seq followed the Zipf's law with IGH disease clones easily identified as outliers. Four hundred and ninety-seven IGH disease clones (median 2, range 0-7 clones/patient) are identified in 90.3% of patients. High hyperdiploid patients have the most IGH disease clones (median 3) while DUX4 subtype has the least (median 1) due to the rearrangements involving the IGH locus. In all, 90.8% of IGH disease clones found by Sanger sequencing are also identified by RNA-Seq. In addition, RNA-Seq identified 43% more IGH disease clones. In 69 patients lacking sensitive IGH targets, targeted NGS IGH MRD showed high correlation (R = 0.93; P = 1.3 × 10-14), better relapse prediction than conventional RQ-PCR MRD using non-IGH targets. In conclusion, RNA-Seq can identify patient-specific clonal IGH junctional sequences for MRD monitoring, adding to its usefulness for molecular diagnosis in childhood B-ALL.
中文翻译:

使用RNA-Seq鉴定IGH疾病克隆以监测儿童B细胞急性淋巴细胞白血病的MRD。
识别患者特异性的克隆IGH / TCR连接序列对于最小化残留疾病(MRD)监测至关重要。常规上,这些连接序列是使用费力的Sanger测序方法对切下的异源双链带进行鉴定的。我们发现IGH在使用RNA-Seq的诊断性B细胞急性淋巴细胞白血病(B-ALL)样本中高表达。因此,我们使用RNA-Seq识别了258个儿童B-ALL样本中的IGH疾病克隆序列,以进行MRD监测。RNA-Seq揭示的背景IGH重排量遵循Zipf定律,其中IGH疾病克隆很容易被识别为异常值。在90.3%的患者中鉴定出479个IGH疾病克隆(中位值为2,范围为0-7个克隆/患者)。由于涉及IGH基因座的重排,高二倍体患者的IGH疾病克隆最多(中位数3),而DUX4亚型的克隆最少(中位数1)。总共,通过Sanger测序发现的90.8%IGH疾病克隆也通过RNA-Seq鉴定。此外,RNA-Seq鉴定出的IGH疾病克隆多出43%。在69位缺乏敏感IGH目标的患者中,靶向NGS IGH MRD表现出高度相关性(R = 0.93; P = 1.3×10-14),比使用非IGH目标的常规RQ-PCR MRD更好的复发预测。总之,RNA-Seq可以识别患者特异性的克隆IGH连接序列以监测MRD,从而增加其在儿童B-ALL分子诊断中的有用性。此外,RNA-Seq鉴定出的IGH疾病克隆多出43%。在69位缺乏敏感IGH目标的患者中,靶向NGS IGH MRD表现出高度相关性(R = 0.93; P = 1.3×10-14),比使用非IGH目标的常规RQ-PCR MRD更好的复发预测。总之,RNA-Seq可以识别患者特异性的克隆IGH连接序列以监测MRD,从而增加其在儿童B-ALL分子诊断中的有用性。此外,RNA-Seq鉴定出的IGH疾病克隆多出43%。在69位缺乏敏感IGH目标的患者中,靶向NGS IGH MRD表现出高度相关性(R = 0.93; P = 1.3×10-14),比使用非IGH目标的常规RQ-PCR MRD更好的复发预测。总之,RNA-Seq可以识别患者特异性的克隆IGH连接序列以监测MRD,从而增加其在儿童B-ALL分子诊断中的有用性。
更新日期:2020-02-25
中文翻译:

使用RNA-Seq鉴定IGH疾病克隆以监测儿童B细胞急性淋巴细胞白血病的MRD。
识别患者特异性的克隆IGH / TCR连接序列对于最小化残留疾病(MRD)监测至关重要。常规上,这些连接序列是使用费力的Sanger测序方法对切下的异源双链带进行鉴定的。我们发现IGH在使用RNA-Seq的诊断性B细胞急性淋巴细胞白血病(B-ALL)样本中高表达。因此,我们使用RNA-Seq识别了258个儿童B-ALL样本中的IGH疾病克隆序列,以进行MRD监测。RNA-Seq揭示的背景IGH重排量遵循Zipf定律,其中IGH疾病克隆很容易被识别为异常值。在90.3%的患者中鉴定出479个IGH疾病克隆(中位值为2,范围为0-7个克隆/患者)。由于涉及IGH基因座的重排,高二倍体患者的IGH疾病克隆最多(中位数3),而DUX4亚型的克隆最少(中位数1)。总共,通过Sanger测序发现的90.8%IGH疾病克隆也通过RNA-Seq鉴定。此外,RNA-Seq鉴定出的IGH疾病克隆多出43%。在69位缺乏敏感IGH目标的患者中,靶向NGS IGH MRD表现出高度相关性(R = 0.93; P = 1.3×10-14),比使用非IGH目标的常规RQ-PCR MRD更好的复发预测。总之,RNA-Seq可以识别患者特异性的克隆IGH连接序列以监测MRD,从而增加其在儿童B-ALL分子诊断中的有用性。此外,RNA-Seq鉴定出的IGH疾病克隆多出43%。在69位缺乏敏感IGH目标的患者中,靶向NGS IGH MRD表现出高度相关性(R = 0.93; P = 1.3×10-14),比使用非IGH目标的常规RQ-PCR MRD更好的复发预测。总之,RNA-Seq可以识别患者特异性的克隆IGH连接序列以监测MRD,从而增加其在儿童B-ALL分子诊断中的有用性。此外,RNA-Seq鉴定出的IGH疾病克隆多出43%。在69位缺乏敏感IGH目标的患者中,靶向NGS IGH MRD表现出高度相关性(R = 0.93; P = 1.3×10-14),比使用非IGH目标的常规RQ-PCR MRD更好的复发预测。总之,RNA-Seq可以识别患者特异性的克隆IGH连接序列以监测MRD,从而增加其在儿童B-ALL分子诊断中的有用性。









































 京公网安备 11010802027423号
京公网安备 11010802027423号