当前位置:
X-MOL 学术
›
J. Proteome Res.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
A Proteogenomic Resource Enabling Integrated Analysis of Listeria Genotype-Proteotype-Phenotype Relationships.
Journal of Proteome Research ( IF 3.8 ) Pub Date : 2020-02-24 , DOI: 10.1021/acs.jproteome.9b00842 Adithi R Varadarajan 1, 2, 3 , Sandra Goetze 1, 3, 4 , Maria P Pavlou 1, 4 , Virginie Grosboillot 1, 5 , Yang Shen 1, 5 , Martin J Loessner 1, 5 , Christian H Ahrens 2, 3 , Bernd Wollscheid 1, 3, 4
Journal of Proteome Research ( IF 3.8 ) Pub Date : 2020-02-24 , DOI: 10.1021/acs.jproteome.9b00842 Adithi R Varadarajan 1, 2, 3 , Sandra Goetze 1, 3, 4 , Maria P Pavlou 1, 4 , Virginie Grosboillot 1, 5 , Yang Shen 1, 5 , Martin J Loessner 1, 5 , Christian H Ahrens 2, 3 , Bernd Wollscheid 1, 3, 4
Affiliation
|
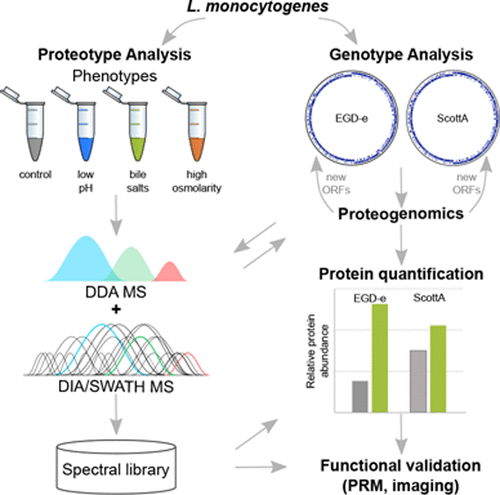
Listeria monocytogenes is an opportunistic foodborne pathogen responsible for listeriosis, a potentially fatal foodborne disease. Many different Listeria strains and serotypes exist, but a proteogenomic resource that bridges the gap in our molecular understanding of the relationships between the Listeria genotypes and phenotypes via proteotypes is still missing. Here, we devised a next-generation proteogenomics strategy that enables the community to rapidly proteotype Listeria strains and relate this information back to the genotype. Based on sequencing and de novo assembly of the two most commonly used Listeria model strains, EGD-e and ScottA, we established two comprehensive Listeria proteogenomic databases. A genome comparison established core- and strain-specific genes potentially responsible for virulence differences. Next, we established a DIA/SWATH-based proteotyping strategy, including a new and robust sample preparation workflow, that enables the reproducible, sensitive, and relative quantitative measurement of Listeria proteotypes. This reusable and publicly available DIA/SWATH library covers 70% of open reading frames of Listeria and represents the most extensive spectral library for Listeria proteotype analysis to date. We used these two new resources to investigate the Listeria proteotype in states mimicking the upper gastrointestinal passage. Exposure of Listeria to bile salts at 37 °C, which simulates conditions encountered in the duodenum, showed significant proteotype perturbations including an increase of FlaA, the structural protein of flagella. Given that Listeria is known to lose its flagella above 30 °C, this was an unexpected finding. The formation of flagella, which might have implications on infectivity, was validated by parallel reaction monitoring and light and scanning electron microscopy. flaA transcript levels did not change significantly upon exposure to bile salts at 37 °C, suggesting regulation at the post-transcriptional level. Together, these analyses provide a comprehensive proteogenomic resource and toolbox for the Listeria community enabling the analysis of Listeria genotype–proteotype–phenotype relationships.
中文翻译:

一种蛋白质组学资源,可对李斯特菌基因型-蛋白质型-表型关系进行综合分析。
单核细胞增生性李斯特菌是导致李斯特菌病(一种潜在的致命食源性疾病)的机会性食源性病原体。存在许多不同的李斯特菌菌株和血清型,但是仍然缺少一种蛋白质组学资源,该资源弥补了我们对通过李斯特菌基因型和表型之间通过蛋白质型之间的关系的分子理解的差距。在这里,我们设计了下一代蛋白质组学策略,使社区能够快速对李斯特菌菌株进行蛋白化,并将此信息与基因型相关联。基于两种最常用的李斯特菌模型菌株EGD-e和ScottA的测序和从头组装,我们建立了两种李斯特菌蛋白质组数据库。基因组比较确定了可能导致毒力差异的核心和菌株特异性基因。接下来,我们建立了一个基于DIA / SWATH的蛋白质分型策略,包括一个新的强大的样品前处理工作流程,该方法可以实现李斯特菌蛋白型的可再现,敏感和相对定量测量。这个可重用且公开可用的DIA / SWATH库涵盖了李斯特菌开放阅读框的70%,代表了迄今为止用于李斯特菌蛋白型分析的最广泛的光谱库。我们使用这两种新资源来研究模拟上胃肠道的州中的李斯特菌蛋白型。接触李斯特菌在37°C的胆汁盐溶液中模拟十二指肠遇到的情况,显示出明显的蛋白型扰动,包括鞭毛的结构蛋白FlaA的增加。考虑到已知利斯特氏菌在30°C以上会失去鞭毛,这是一个出乎意料的发现。鞭毛的形成可能对传染性有影响,已通过平行反应监测以及光和扫描电子显微镜进行了验证。暴露于37°C的胆汁盐后,flaA转录水平没有明显变化,表明在转录后水平有调控。这些分析共同为李斯特菌属提供了全面的蛋白质组学资源和工具箱,从而可以对李斯特菌进行分析 基因型-蛋白型-表型的关系。
更新日期:2020-02-24
中文翻译:

一种蛋白质组学资源,可对李斯特菌基因型-蛋白质型-表型关系进行综合分析。
单核细胞增生性李斯特菌是导致李斯特菌病(一种潜在的致命食源性疾病)的机会性食源性病原体。存在许多不同的李斯特菌菌株和血清型,但是仍然缺少一种蛋白质组学资源,该资源弥补了我们对通过李斯特菌基因型和表型之间通过蛋白质型之间的关系的分子理解的差距。在这里,我们设计了下一代蛋白质组学策略,使社区能够快速对李斯特菌菌株进行蛋白化,并将此信息与基因型相关联。基于两种最常用的李斯特菌模型菌株EGD-e和ScottA的测序和从头组装,我们建立了两种李斯特菌蛋白质组数据库。基因组比较确定了可能导致毒力差异的核心和菌株特异性基因。接下来,我们建立了一个基于DIA / SWATH的蛋白质分型策略,包括一个新的强大的样品前处理工作流程,该方法可以实现李斯特菌蛋白型的可再现,敏感和相对定量测量。这个可重用且公开可用的DIA / SWATH库涵盖了李斯特菌开放阅读框的70%,代表了迄今为止用于李斯特菌蛋白型分析的最广泛的光谱库。我们使用这两种新资源来研究模拟上胃肠道的州中的李斯特菌蛋白型。接触李斯特菌在37°C的胆汁盐溶液中模拟十二指肠遇到的情况,显示出明显的蛋白型扰动,包括鞭毛的结构蛋白FlaA的增加。考虑到已知利斯特氏菌在30°C以上会失去鞭毛,这是一个出乎意料的发现。鞭毛的形成可能对传染性有影响,已通过平行反应监测以及光和扫描电子显微镜进行了验证。暴露于37°C的胆汁盐后,flaA转录水平没有明显变化,表明在转录后水平有调控。这些分析共同为李斯特菌属提供了全面的蛋白质组学资源和工具箱,从而可以对李斯特菌进行分析 基因型-蛋白型-表型的关系。











































 京公网安备 11010802027423号
京公网安备 11010802027423号