当前位置:
X-MOL 学术
›
J. Proteome Res.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Hat1-Dependent Lysine Acetylation Targets Diverse Cellular Functions.
Journal of Proteome Research ( IF 3.8 ) Pub Date : 2020-02-21 , DOI: 10.1021/acs.jproteome.9b00843 Paula A Agudelo Garcia 1 , Prabakaran Nagarajan 1 , Mark R Parthun 1
Journal of Proteome Research ( IF 3.8 ) Pub Date : 2020-02-21 , DOI: 10.1021/acs.jproteome.9b00843 Paula A Agudelo Garcia 1 , Prabakaran Nagarajan 1 , Mark R Parthun 1
Affiliation
|
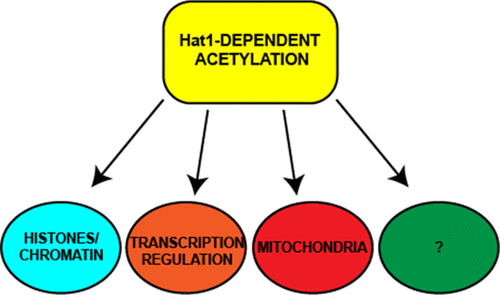
Lysine acetylation has emerged as one of the most important post-translational modifications, regulating different biological processes. However, its regulation by lysine acetyltransferases is still unclear in most cases. Hat1 is a lysine acetyltransferase originally identified based on its ability to acetylate histones. Using an unbiased proteomics approach, we have determined how loss of Hat1 affects the mammalian acetylome. Hat1+/+ and Hat1-/- mouse embryonic fibroblast cell lines were grown in both glucose- and galactose-containing media, as Hat1 is required for growth on galactose, and Hat1-/- cells exhibit defects in mitochondrial function. Following trypsin digestion of whole cell extracts, acetylated peptides were enriched by acetyllysine affinity purification, and acetylated peptides were identified and analyzed by label-free quantitation. Comparison of the acetylome from Hat1+/+ cells grown on galactose and glucose demonstrated that there are large carbon source-dependent changes in the mammalian acetylome where the acetylation of enzymes involved in glycolysis were the most affected. Comparisons of the acetylomes from Hat1+/+ and Hat1-/- cells identified 65 proteins whose acetylation decreased by at least 2.5-fold in cells lacking Hat1. In Hat1-/- cells, acetylation of the autoregulatory loop of CBP (CREB-binding protein) was the most highly affected, decreasing by up to 20-fold. In addition to the proteins involved in chromatin structure, Hat1-dependent acetylation was also found in a number of transcriptional regulators, including p53 and mitochondrial proteins. Hat1 mitochondrial localization suggests that it may be directly involved in the acetylation of mitochondrial proteins. Data are available via ProteomeXchange with identifier PXD017362.
中文翻译:

依赖于Hat1的赖氨酸乙酰化靶向多种细胞功能。
赖氨酸乙酰化已成为最重要的翻译后修饰之一,调节着不同的生物学过程。然而,在大多数情况下,赖氨酸乙酰基转移酶对其调节的作用仍不清楚。Hat1是赖氨酸乙酰基转移酶,最初是基于其乙酰化组蛋白的能力而鉴定的。使用无偏蛋白质组学方法,我们已经确定Hat1的丧失如何影响哺乳动物的乙酰基。Hat1 + / +和Hat1-/-小鼠胚胎成纤维细胞系在含葡萄糖和半乳糖的培养基中生长,因为Hat1在半乳糖上生长是必需的,并且Hat1-/-细胞在线粒体功能方面表现出缺陷。用胰蛋白酶消化全细胞提取物后,通过乙酰赖氨酸亲和纯化来富集乙酰化的肽,然后通过无标记定量来鉴定和分析乙酰化的肽。比较半乳糖和葡萄糖上生长的Hat1 + / +细胞中的乙酰酶组,发现哺乳动物的乙酰酶组中碳源依赖性变化较大,其中糖酵解酶的乙酰化受影响最大。对来自Hat1 + / +和Hat1-/-细胞的乙酰基的比较发现,在缺乏Hat1的细胞中,有65种蛋白质的乙酰化程度降低了至少2.5倍。在Hat1-/-细胞中,CBP(CREB结合蛋白)的自调节环的乙酰化受到的影响最大,最多降低20倍。除了涉及染色质结构的蛋白质外,还在许多转录调节因子(包括p53和线粒体蛋白)中发现了Hat1依赖性乙酰化。Hat1线粒体定位表明它可能直接参与线粒体蛋白的乙酰化。数据可通过ProteomeXchange获得,其标识符为PXD017362。
更新日期:2020-02-21
中文翻译:

依赖于Hat1的赖氨酸乙酰化靶向多种细胞功能。
赖氨酸乙酰化已成为最重要的翻译后修饰之一,调节着不同的生物学过程。然而,在大多数情况下,赖氨酸乙酰基转移酶对其调节的作用仍不清楚。Hat1是赖氨酸乙酰基转移酶,最初是基于其乙酰化组蛋白的能力而鉴定的。使用无偏蛋白质组学方法,我们已经确定Hat1的丧失如何影响哺乳动物的乙酰基。Hat1 + / +和Hat1-/-小鼠胚胎成纤维细胞系在含葡萄糖和半乳糖的培养基中生长,因为Hat1在半乳糖上生长是必需的,并且Hat1-/-细胞在线粒体功能方面表现出缺陷。用胰蛋白酶消化全细胞提取物后,通过乙酰赖氨酸亲和纯化来富集乙酰化的肽,然后通过无标记定量来鉴定和分析乙酰化的肽。比较半乳糖和葡萄糖上生长的Hat1 + / +细胞中的乙酰酶组,发现哺乳动物的乙酰酶组中碳源依赖性变化较大,其中糖酵解酶的乙酰化受影响最大。对来自Hat1 + / +和Hat1-/-细胞的乙酰基的比较发现,在缺乏Hat1的细胞中,有65种蛋白质的乙酰化程度降低了至少2.5倍。在Hat1-/-细胞中,CBP(CREB结合蛋白)的自调节环的乙酰化受到的影响最大,最多降低20倍。除了涉及染色质结构的蛋白质外,还在许多转录调节因子(包括p53和线粒体蛋白)中发现了Hat1依赖性乙酰化。Hat1线粒体定位表明它可能直接参与线粒体蛋白的乙酰化。数据可通过ProteomeXchange获得,其标识符为PXD017362。











































 京公网安备 11010802027423号
京公网安备 11010802027423号