当前位置:
X-MOL 学术
›
Eur. J. Hum. Genet.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Incorporating information from markers in LD with test locus for detecting imprinting and maternal effects.
European Journal of Human Genetics ( IF 3.7 ) Pub Date : 2020-02-20 , DOI: 10.1038/s41431-020-0590-3 Fangyuan Zhang 1 , Shili Lin 2
European Journal of Human Genetics ( IF 3.7 ) Pub Date : 2020-02-20 , DOI: 10.1038/s41431-020-0590-3 Fangyuan Zhang 1 , Shili Lin 2
Affiliation
|
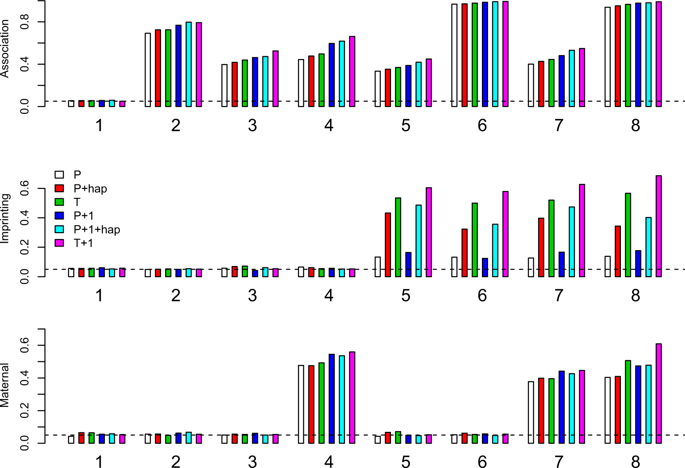
Numerous statistical methods have been developed to explore genomic imprinting and maternal effects by identifying parent-of-origin patterns in complex human diseases. However, because most of these methods only use available locus-specific genotype data, it is sometimes impossible for them to infer the distribution of parental origin of a variant allele, especially when some genotypes are missing. In this article, we propose a two-step approach, LIMEhap, to improve upon a recent partial likelihood inference method. In the first step, the distribution of the missing genotypes is inferred through the construction of haplotypes by using information from nearby loci. In the second step, a partial likelihood method is applied to the inferred data. To substantiate the validity of the proposed procedures, we simulated data in a genomic region of gene GPX1. The results show that, by borrowing genetic information from nearby loci, the power of the proposed method can be close to that with complete genotype data at the locus of interest. Since the inference on the genotype distribution is made under the assumption of Hardy-Weinberg Equilibrium (HWE), we further studied the robustness of LIMEhap to violation of HWE. Finally, we demonstrate the utility of LIMEhap by applying it to an autism dataset.
中文翻译:

将来自 LD 中标记的信息与测试位点相结合,以检测印记和母体效应。
已经开发了许多统计方法来通过识别复杂人类疾病中的亲本模式来探索基因组印记和母体效应。然而,由于这些方法中的大多数只使用可用的基因座特异性基因型数据,有时它们无法推断变异等位基因的亲本来源的分布,尤其是当某些基因型缺失时。在本文中,我们提出了一种两步法 LIMEhap,以改进最近的部分似然推理方法。在第一步中,通过使用来自附近基因座的信息构建单倍型来推断缺失基因型的分布。第二步,对推断数据应用部分似然方法。为证实拟议程序的有效性,我们模拟了基因 GPX1 基因组区域中的数据。结果表明,通过从附近基因座借用遗传信息,所提出方法的功效可以接近感兴趣基因座的完整基因型数据。由于对基因型分布的推断是在 Hardy-Weinberg Equilibrium (HWE) 的假设下进行的,我们进一步研究了 LIMEhap 对违反 HWE 的鲁棒性。最后,我们通过将 LIMEhap 应用于自闭症数据集来证明其实用性。我们进一步研究了 LIMEhap 对违反 HWE 的鲁棒性。最后,我们通过将 LIMEhap 应用于自闭症数据集来证明其实用性。我们进一步研究了 LIMEhap 对违反 HWE 的鲁棒性。最后,我们通过将 LIMEhap 应用于自闭症数据集来证明其实用性。
更新日期:2020-02-20
中文翻译:

将来自 LD 中标记的信息与测试位点相结合,以检测印记和母体效应。
已经开发了许多统计方法来通过识别复杂人类疾病中的亲本模式来探索基因组印记和母体效应。然而,由于这些方法中的大多数只使用可用的基因座特异性基因型数据,有时它们无法推断变异等位基因的亲本来源的分布,尤其是当某些基因型缺失时。在本文中,我们提出了一种两步法 LIMEhap,以改进最近的部分似然推理方法。在第一步中,通过使用来自附近基因座的信息构建单倍型来推断缺失基因型的分布。第二步,对推断数据应用部分似然方法。为证实拟议程序的有效性,我们模拟了基因 GPX1 基因组区域中的数据。结果表明,通过从附近基因座借用遗传信息,所提出方法的功效可以接近感兴趣基因座的完整基因型数据。由于对基因型分布的推断是在 Hardy-Weinberg Equilibrium (HWE) 的假设下进行的,我们进一步研究了 LIMEhap 对违反 HWE 的鲁棒性。最后,我们通过将 LIMEhap 应用于自闭症数据集来证明其实用性。我们进一步研究了 LIMEhap 对违反 HWE 的鲁棒性。最后,我们通过将 LIMEhap 应用于自闭症数据集来证明其实用性。我们进一步研究了 LIMEhap 对违反 HWE 的鲁棒性。最后,我们通过将 LIMEhap 应用于自闭症数据集来证明其实用性。











































 京公网安备 11010802027423号
京公网安备 11010802027423号