Molecular Metabolism ( IF 8.1 ) Pub Date : 2020-02-14 , DOI: 10.1016/j.molmet.2020.01.017 Verena M Prade 1 , Thomas Kunzke 1 , Annette Feuchtinger 1 , Maria Rohm 2 , Birgit Luber 3 , Florian Lordick 4 , Achim Buck 1 , Axel Walch 1
|
Background
Imaging mass spectrometry enables in situ label-free detection of thousands of metabolites from intact tissue samples. However, automated steps for multi-omics analyses and interpretation of histological images have not yet been implemented in mass spectrometry data analysis workflows. The characterization of molecular properties within cellular and histological features is done via time-consuming, non-objective, and irreproducible definitions of regions of interest, which are often accompanied by a loss of spatial resolution due to mass spectra averaging.
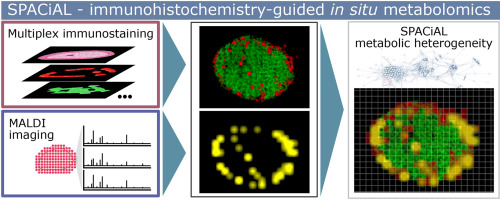
Methods: We developed a new imaging pipeline called Spatial Correlation Image Analysis (SPACiAL), which is a computational multimodal workflow designed to combine molecular imaging data with multiplex immunohistochemistry (IHC). SPACiAL allows comprehensive and spatially resolved in situ correlation analyses on a cellular resolution. To demonstrate the method, matrix-assisted laser desorption-ionization (MALDI) Fourier-transform ion cyclotron resonance (FTICR) imaging mass spectrometry of metabolites and multiplex IHC staining were performed on the very same tissue section of mouse pancreatic islets and on human gastric cancer tissue specimens. The SPACiAL pipeline was used to perform an automatic, semantic-based, functional tissue annotation of histological and cellular features to identify metabolic profiles. Spatial correlation networks were generated to analyze metabolic heterogeneity associated with cellular features.
Results: To demonstrate the new method, the SPACiAL pipeline was used to identify metabolic signatures of alpha and beta cells within islets of Langerhans, which are cell types that are not distinguishable via morphology alone. The semantic-based, functional tissue annotation allows an unprecedented analysis of metabolic heterogeneity via the generation of spatial correlation networks. Additionally, we demonstrated intra- and intertumoral metabolic heterogeneity within HER2/neu-positive and -negative gastric tumor cells.
Conclusions: We developed the SPACiAL workflow to provide IHC-guided in situ metabolomics on intact tissue sections. Diminishing the workload by automated recognition of histological and functional features, the pipeline allows comprehensive analyses of metabolic heterogeneity. The multimodality of immunohistochemical staining and extensive molecular information from imaging mass spectrometry has the advantage of increasing both the efficiency and precision for spatially resolved analyses of specific cell types. The SPACiAL method is a stepping stone for the objective analysis of high-throughput, multi-omics data from clinical research and practice that is required for diagnostics, biomarker discovery, or therapy response prediction.
中文翻译:

从头到尾发现免疫异型指导成像质谱的代谢异质性。
背景
成像质谱能够从完整的组织样本中无标记地原位检测数千种代谢产物。但是,质谱数据分析工作流程中尚未实现用于多组学分析和组织学图像解释的自动化步骤。细胞和组织学特征内分子特性的表征是通过耗时,非客观且不可重现的目标区域定义来完成的,通常由于质谱平均而伴随着空间分辨率的下降。
方法:我们开发了一种称为空间相关图像分析(SPACiAL)的新成像管道,该管道是一种计算性多模式工作流程,旨在将分子成像数据与多重免疫组织化学(IHC)结合在一起。SPACiAL允许在原位进行全面的空间解析细胞分辨率的相关性分析。为了证明该方法,对小鼠胰岛和人体胃癌的同一组织切片进行了代谢物的基质辅助激光解吸电离(MALDI)傅里叶变换离子回旋共振(FTICR)成像质谱和多重IHC染色组织标本。SPACiAL管道用于对组织学和细胞特征执行基于语义的自动功能组织注释,以识别代谢谱。生成空间相关网络以分析与细胞特征相关的代谢异质性。
结果:为了证明该新方法,使用了SPACiAL管道来识别Langerhans胰岛中的α和β细胞的代谢特征,这是不能单独通过形态学区分的细胞类型。基于语义的功能组织注释可通过生成空间相关网络对代谢异质性进行前所未有的分析。此外,我们证明了HER2 / neu阳性和阴性胃肿瘤细胞内和瘤内代谢异质性。
结论:我们开发了SPACiAL工作流程,以在完整组织切片上提供IHC指导的原位代谢组学。通过自动识别组织学和功能特征,减少了工作量,该管道可以对代谢异质性进行全面分析。免疫组织化学染色的多态性和来自成像质谱的大量分子信息具有提高特定细胞类型的空间分辨分析效率和精度的优势。SPACiAL方法是诊断,生物标志物发现或治疗反应预测所需的来自临床研究和实践的高通量,多组学数据的客观分析的垫脚石。



























 京公网安备 11010802027423号
京公网安备 11010802027423号