Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
A Quantitative Genetic Interaction Map of HIV Infection.
Molecular Cell ( IF 14.5 ) Pub Date : 2020-02-20 , DOI: 10.1016/j.molcel.2020.02.004 David E Gordon 1 , Ariane Watson 2 , Assen Roguev 3 , Simin Zheng 4 , Gwendolyn M Jang 3 , Joshua Kane 3 , Jiewei Xu 3 , Jeffrey Z Guo 3 , Erica Stevenson 1 , Danielle L Swaney 1 , Kathy Franks-Skiba 3 , Erik Verschueren 5 , Michael Shales 3 , David C Crosby 6 , Alan D Frankel 6 , Alexander Marson 7 , Ivan Marazzi 4 , Gerard Cagney 2 , Nevan J Krogan 8
Molecular Cell ( IF 14.5 ) Pub Date : 2020-02-20 , DOI: 10.1016/j.molcel.2020.02.004 David E Gordon 1 , Ariane Watson 2 , Assen Roguev 3 , Simin Zheng 4 , Gwendolyn M Jang 3 , Joshua Kane 3 , Jiewei Xu 3 , Jeffrey Z Guo 3 , Erica Stevenson 1 , Danielle L Swaney 1 , Kathy Franks-Skiba 3 , Erik Verschueren 5 , Michael Shales 3 , David C Crosby 6 , Alan D Frankel 6 , Alexander Marson 7 , Ivan Marazzi 4 , Gerard Cagney 2 , Nevan J Krogan 8
Affiliation
|
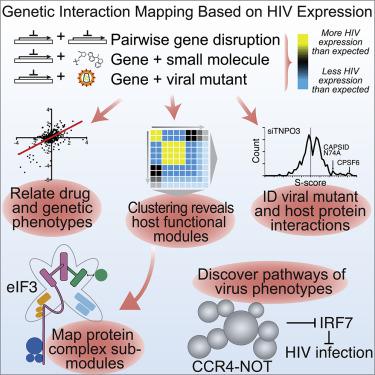
We have developed a platform for quantitative genetic interaction mapping using viral infectivity as a functional readout and constructed a viral host-dependency epistasis map (vE-MAP) of 356 human genes linked to HIV function, comprising >63,000 pairwise genetic perturbations. The vE-MAP provides an expansive view of the genetic dependencies underlying HIV infection and can be used to identify drug targets and study viral mutations. We found that the RNA deadenylase complex, CNOT, is a central player in the vE-MAP and show that knockout of CNOT1, 10, and 11 suppressed HIV infection in primary T cells by upregulating innate immunity pathways. This phenotype was rescued by deletion of IRF7, a transcription factor regulating interferon-stimulated genes, revealing a previously unrecognized host signaling pathway involved in HIV infection. The vE-MAP represents a generic platform that can be used to study the global effects of how different pathogens hijack and rewire the host during infection.
中文翻译:

HIV感染的定量遗传相互作用图。
我们已经开发了使用病毒感染性作为功能读数进行定量遗传相互作用作图的平台,并构建了与HIV功能相关的356个人类基因的病毒宿主依赖性上位图(vE-MAP),包括> 63,000个成对的遗传扰动。vE-MAP提供了有关HIV感染的遗传依赖性的广泛视图,可用于识别药物靶标和研究病毒突变。我们发现,RNA腺苷酸酶复合物CNOT是vE-MAP的核心角色,并显示CNOT1、10和11的敲除通过上调先天免疫途径抑制了原代T细胞中的HIV感染。该表型通过删除IRF7(一种调节干扰素刺激基因的转录因子)而得以挽救,揭示了先前未认识到的参与HIV感染的宿主信号通路。
更新日期:2020-02-20
中文翻译:

HIV感染的定量遗传相互作用图。
我们已经开发了使用病毒感染性作为功能读数进行定量遗传相互作用作图的平台,并构建了与HIV功能相关的356个人类基因的病毒宿主依赖性上位图(vE-MAP),包括> 63,000个成对的遗传扰动。vE-MAP提供了有关HIV感染的遗传依赖性的广泛视图,可用于识别药物靶标和研究病毒突变。我们发现,RNA腺苷酸酶复合物CNOT是vE-MAP的核心角色,并显示CNOT1、10和11的敲除通过上调先天免疫途径抑制了原代T细胞中的HIV感染。该表型通过删除IRF7(一种调节干扰素刺激基因的转录因子)而得以挽救,揭示了先前未认识到的参与HIV感染的宿主信号通路。










































 京公网安备 11010802027423号
京公网安备 11010802027423号