当前位置:
X-MOL 学术
›
ACS Comb. Sci.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
PTML Model of ChEMBL Compounds Assays for Vitamin Derivatives.
ACS Combinatorial Science Pub Date : 2020-02-13 , DOI: 10.1021/acscombsci.9b00166 Ricardo Santana 1, 2 , Robin Zuluaga 3 , Piedad Gañán 4 , Sonia Arrasate 5 , Enrique Onieva Caracuel 1 , Humbert González-Díaz 5, 6
ACS Combinatorial Science Pub Date : 2020-02-13 , DOI: 10.1021/acscombsci.9b00166 Ricardo Santana 1, 2 , Robin Zuluaga 3 , Piedad Gañán 4 , Sonia Arrasate 5 , Enrique Onieva Caracuel 1 , Humbert González-Díaz 5, 6
Affiliation
|
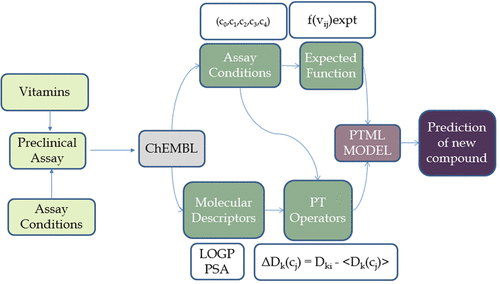
Determining the biological activity of vitamin derivatives is needed given that organic synthesis of analogs of vitamins is an active field of interest for medicinal chemistry, pharmaceuticals, and food additives. Accordingly, scientists from different disciplines perform preclinical assays (nij) with a considerable combination of assay conditions (cj). Indeed, the ChEMBL platform contains a database that includes results from 36 220 different biological activity bioassays of 21 240 different vitamins and vitamin derivatives. These assays present are heterogeneous in terms of assay combinations of cj. They are focused on >500 different biological activity parameters (c0), >340 different targets (c1), >6200 types of cell (c2), >120 organisms of assay (c3), and >60 assay strains (c4). It includes a total of >1850 niacin assays, >1580 tretinoin assays, >1580 retinol assays, 857 ascorbic acid assays, etc. Given the complexity of this combinatorial data in terms of being assimilated by researchers, we propose to build a model by combining perturbation theory (PT) and machine learning (ML). Through this study, we propose a PTML (PT + ML) combinatorial model for ChEMBL results on biological activity of vitamins and vitamins derivatives. The linear discriminant analysis (LDA) model presented the following results for training subset a: specificity (%) = 90.38, sensitivity (%) = 87.51, and accuracy (%) = 89.89. The model showed the following results for the external validation subset: specificity (%) = 90.58, sensitivity (%) = 87.72, and accuracy (%) = 90.09. Different types of linear and nonlinear PTML models, such as logistic regression (LR), classification tree (CT), näive Bayes (NB), and random Forest (RF), were applied to contrast the capacity of prediction. The PTML-LDA model predicts with more accuracy by applying combinatorial descriptors. In addition, a PCA experiment with chemical structure descriptors allowed us to characterize the high structural diversity of the chemical space studied. In any case, PTML models using chemical structure descriptors do not improve the performance of the PTML-LDA model based on ALOGP and PSA. We can conclude that the three variable PTML-LDA model is a simplified and adaptable tool for the prediction, for different experiment combinations, the biological activity of derivative vitamins.
中文翻译:

维生素衍生物的ChEMBL化合物分析的PTML模型。
鉴于维生素类似物的有机合成是药物化学,药物和食品添加剂关注的一个活跃领域,因此需要确定维生素衍生物的生物活性。相应地,来自不同学科的科学家通过大量的检测条件(cj)进行临床前检测(nij)。实际上,ChEMBL平台包含一个数据库,该数据库包含来自21 240种不同维生素和维生素衍生物的36 220种不同生物活性生物测定的结果。存在的这些测定就cj的测定组合而言是异质的。他们专注于> 500种不同的生物活性参数(c0),> 340种不同的靶标(c1),> 6200种类型的细胞(c2),> 120种测定生物(c3)和> 60种测定菌株(c4)。它总共包括> 1850个烟酸测定,1580维甲酸测定,> 1580视黄醇测定,857抗坏血酸测定等。考虑到研究人员对这些组合数据的复杂性,我们建议通过结合摄动理论(PT)和机器学习(ML)来构建模型。通过这项研究,我们提出了ChEMBL结果对维生素和维生素衍生物的生物活性的PTML(PT + ML)组合模型。线性判别分析(LDA)模型为训练子集a提供了以下结果:特异性(%)= 90.38,灵敏度(%)= 87.51,准确性(%)= 89.89。对于外部验证子集,该模型显示以下结果:特异性(%)= 90.58,灵敏度(%)= 87.72,准确性(%)= 90.09。不同类型的线性和非线性PTML模型,例如逻辑回归(LR),分类树(CT),应用朴素贝叶斯(NB)和随机森林(RF)来对比预测能力。PTML-LDA模型通过应用组合描述符来进行更准确的预测。此外,具有化学结构描述符的PCA实验使我们能够表征所研究化学空间的高结构多样性。无论如何,使用化学结构描述符的PTML模型都不会提高基于ALOGP和PSA的PTML-LDA模型的性能。我们可以得出结论,三变量PTML-LDA模型是一种简化且适应性强的工具,用于针对不同的实验组合预测衍生维生素的生物活性。具有化学结构描述符的PCA实验使我们能够表征所研究化学空间的高结构多样性。无论如何,使用化学结构描述符的PTML模型都不会提高基于ALOGP和PSA的PTML-LDA模型的性能。我们可以得出结论,三变量PTML-LDA模型是一种简化且适应性强的工具,用于针对不同的实验组合预测衍生维生素的生物活性。具有化学结构描述符的PCA实验使我们能够表征所研究化学空间的高结构多样性。无论如何,使用化学结构描述符的PTML模型都不会提高基于ALOGP和PSA的PTML-LDA模型的性能。我们可以得出结论,三变量PTML-LDA模型是一种简化且适应性强的工具,用于针对不同的实验组合预测衍生维生素的生物活性。
更新日期:2020-02-13
中文翻译:

维生素衍生物的ChEMBL化合物分析的PTML模型。
鉴于维生素类似物的有机合成是药物化学,药物和食品添加剂关注的一个活跃领域,因此需要确定维生素衍生物的生物活性。相应地,来自不同学科的科学家通过大量的检测条件(cj)进行临床前检测(nij)。实际上,ChEMBL平台包含一个数据库,该数据库包含来自21 240种不同维生素和维生素衍生物的36 220种不同生物活性生物测定的结果。存在的这些测定就cj的测定组合而言是异质的。他们专注于> 500种不同的生物活性参数(c0),> 340种不同的靶标(c1),> 6200种类型的细胞(c2),> 120种测定生物(c3)和> 60种测定菌株(c4)。它总共包括> 1850个烟酸测定,1580维甲酸测定,> 1580视黄醇测定,857抗坏血酸测定等。考虑到研究人员对这些组合数据的复杂性,我们建议通过结合摄动理论(PT)和机器学习(ML)来构建模型。通过这项研究,我们提出了ChEMBL结果对维生素和维生素衍生物的生物活性的PTML(PT + ML)组合模型。线性判别分析(LDA)模型为训练子集a提供了以下结果:特异性(%)= 90.38,灵敏度(%)= 87.51,准确性(%)= 89.89。对于外部验证子集,该模型显示以下结果:特异性(%)= 90.58,灵敏度(%)= 87.72,准确性(%)= 90.09。不同类型的线性和非线性PTML模型,例如逻辑回归(LR),分类树(CT),应用朴素贝叶斯(NB)和随机森林(RF)来对比预测能力。PTML-LDA模型通过应用组合描述符来进行更准确的预测。此外,具有化学结构描述符的PCA实验使我们能够表征所研究化学空间的高结构多样性。无论如何,使用化学结构描述符的PTML模型都不会提高基于ALOGP和PSA的PTML-LDA模型的性能。我们可以得出结论,三变量PTML-LDA模型是一种简化且适应性强的工具,用于针对不同的实验组合预测衍生维生素的生物活性。具有化学结构描述符的PCA实验使我们能够表征所研究化学空间的高结构多样性。无论如何,使用化学结构描述符的PTML模型都不会提高基于ALOGP和PSA的PTML-LDA模型的性能。我们可以得出结论,三变量PTML-LDA模型是一种简化且适应性强的工具,用于针对不同的实验组合预测衍生维生素的生物活性。具有化学结构描述符的PCA实验使我们能够表征所研究化学空间的高结构多样性。无论如何,使用化学结构描述符的PTML模型都不会提高基于ALOGP和PSA的PTML-LDA模型的性能。我们可以得出结论,三变量PTML-LDA模型是一种简化且适应性强的工具,用于针对不同的实验组合预测衍生维生素的生物活性。









































 京公网安备 11010802027423号
京公网安备 11010802027423号