当前位置:
X-MOL 学术
›
npj Genom. Med.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Allele-specific miRNA-binding analysis identifies candidate target genes for breast cancer risk.
npj Genomic Medicine ( IF 4.7 ) Pub Date : 2020-02-13 , DOI: 10.1038/s41525-019-0112-9 Ana Jacinta-Fernandes 1, 2, 3 , Joana M Xavier 1, 2, 3 , Ramiro Magno 2, 3 , Joel G Lage 1, 2, 3 , Ana-Teresa Maia 1, 2, 3
npj Genomic Medicine ( IF 4.7 ) Pub Date : 2020-02-13 , DOI: 10.1038/s41525-019-0112-9 Ana Jacinta-Fernandes 1, 2, 3 , Joana M Xavier 1, 2, 3 , Ramiro Magno 2, 3 , Joel G Lage 1, 2, 3 , Ana-Teresa Maia 1, 2, 3
Affiliation
|
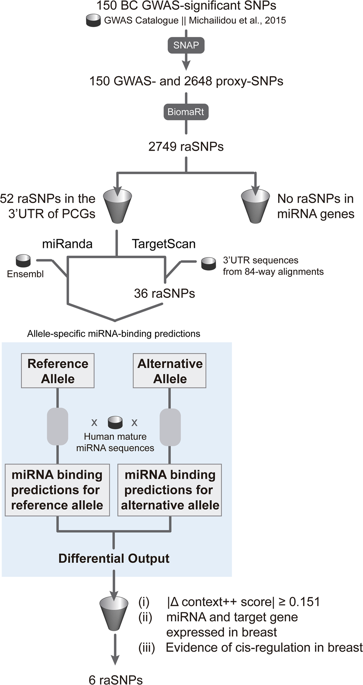
Most breast cancer (BC) risk-associated single-nucleotide polymorphisms (raSNPs) identified in genome-wide association studies (GWAS) are believed to cis-regulate the expression of genes. We hypothesise that cis-regulatory variants contributing to disease risk may be affecting microRNA (miRNA) genes and/or miRNA binding. To test this, we adapted two miRNA-binding prediction algorithms-TargetScan and miRanda-to perform allele-specific queries, and integrated differential allelic expression (DAE) and expression quantitative trait loci (eQTL) data, to query 150 genome-wide significant ( P ≤ 5 × 10 - 8 ) raSNPs, plus proxies. We found that no raSNP mapped to a miRNA gene, suggesting that altered miRNA targeting is an unlikely mechanism involved in BC risk. Also, 11.5% (6 out of 52) raSNPs located in 3'-untranslated regions of putative miRNA target genes were predicted to alter miRNA::mRNA (messenger RNA) pair binding stability in five candidate target genes. Of these, we propose RNF115, at locus 1q21.1, as a strong novel target gene associated with BC risk, and reinforce the role of miRNA-mediated cis-regulation at locus 19p13.11. We believe that integrating allele-specific querying in miRNA-binding prediction, and data supporting cis-regulation of expression, improves the identification of candidate target genes in BC risk, as well as in other common cancers and complex diseases.
中文翻译:

等位基因特异的miRNA结合分析可确定可能罹患乳腺癌的靶基因。
据信在全基因组关联研究(GWAS)中鉴定出的大多数与乳腺癌(BC)风险相关的单核苷酸多态性(raSNPs)均可以顺式调节基因的表达。我们假设顺式调节变异体可能导致疾病风险,可能会影响microRNA(miRNA)基因和/或miRNA结合。为了测试这一点,我们改编了两个miRNA结合预测算法TargetScan和miRanda以执行等位基因特异性查询,并整合了差异等位基因表达(DAE)和表达定量性状基因座(eQTL)数据,以查询150个全基因组显着( P≤5×10-8)raSNP,加上代理。我们发现,没有raSNP定位到miRNA基因,这表明改变miRNA的靶向性是参与BC风险的不太可能的机制。此外,位于3'端的11.5%(52个中的6个)raSNP 预测的miRNA靶基因的非翻译区会改变五个候选靶基因中miRNA :: mRNA(信使RNA)对的结合稳定性。其中,我们建议在基因座1q21.1处的RNF115作为与BC风险相关的强新靶基因,并加强在位点19p13.11处miRNA介导的顺式调节的作用。我们相信,将等位基因特异性查询整合到miRNA结合预测中,以及支持顺式表达调控的数据,可以提高对BC风险以及其他常见癌症和复杂疾病中候选靶基因的识别。并增强19p13.11位点的miRNA介导的顺式调节作用。我们相信,将等位基因特异性查询整合到miRNA结合预测中,以及支持顺式表达调控的数据,可以提高对BC风险以及其他常见癌症和复杂疾病中候选靶基因的识别。并增强19p13.11位点的miRNA介导的顺式调节作用。我们相信,将等位基因特异性查询整合到miRNA结合预测中,以及支持顺式表达调控的数据,可以提高对BC风险以及其他常见癌症和复杂疾病中候选靶基因的识别。
更新日期:2020-02-13
中文翻译:

等位基因特异的miRNA结合分析可确定可能罹患乳腺癌的靶基因。
据信在全基因组关联研究(GWAS)中鉴定出的大多数与乳腺癌(BC)风险相关的单核苷酸多态性(raSNPs)均可以顺式调节基因的表达。我们假设顺式调节变异体可能导致疾病风险,可能会影响microRNA(miRNA)基因和/或miRNA结合。为了测试这一点,我们改编了两个miRNA结合预测算法TargetScan和miRanda以执行等位基因特异性查询,并整合了差异等位基因表达(DAE)和表达定量性状基因座(eQTL)数据,以查询150个全基因组显着( P≤5×10-8)raSNP,加上代理。我们发现,没有raSNP定位到miRNA基因,这表明改变miRNA的靶向性是参与BC风险的不太可能的机制。此外,位于3'端的11.5%(52个中的6个)raSNP 预测的miRNA靶基因的非翻译区会改变五个候选靶基因中miRNA :: mRNA(信使RNA)对的结合稳定性。其中,我们建议在基因座1q21.1处的RNF115作为与BC风险相关的强新靶基因,并加强在位点19p13.11处miRNA介导的顺式调节的作用。我们相信,将等位基因特异性查询整合到miRNA结合预测中,以及支持顺式表达调控的数据,可以提高对BC风险以及其他常见癌症和复杂疾病中候选靶基因的识别。并增强19p13.11位点的miRNA介导的顺式调节作用。我们相信,将等位基因特异性查询整合到miRNA结合预测中,以及支持顺式表达调控的数据,可以提高对BC风险以及其他常见癌症和复杂疾病中候选靶基因的识别。并增强19p13.11位点的miRNA介导的顺式调节作用。我们相信,将等位基因特异性查询整合到miRNA结合预测中,以及支持顺式表达调控的数据,可以提高对BC风险以及其他常见癌症和复杂疾病中候选靶基因的识别。










































 京公网安备 11010802027423号
京公网安备 11010802027423号