Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Mammalian ALKBH1 serves as an N6-mA demethylase of unpairing DNA.
Cell Research ( IF 28.1 ) Pub Date : 2020-02-12 , DOI: 10.1038/s41422-019-0237-5 Min Zhang 1 , Shumin Yang 1 , Raman Nelakanti 2 , Wentao Zhao 1 , Gaochao Liu 1 , Zheng Li 2 , Xiaohui Liu 3 , Tao Wu 2, 4 , Andrew Xiao 2 , Haitao Li 1
Cell Research ( IF 28.1 ) Pub Date : 2020-02-12 , DOI: 10.1038/s41422-019-0237-5 Min Zhang 1 , Shumin Yang 1 , Raman Nelakanti 2 , Wentao Zhao 1 , Gaochao Liu 1 , Zheng Li 2 , Xiaohui Liu 3 , Tao Wu 2, 4 , Andrew Xiao 2 , Haitao Li 1
Affiliation
|
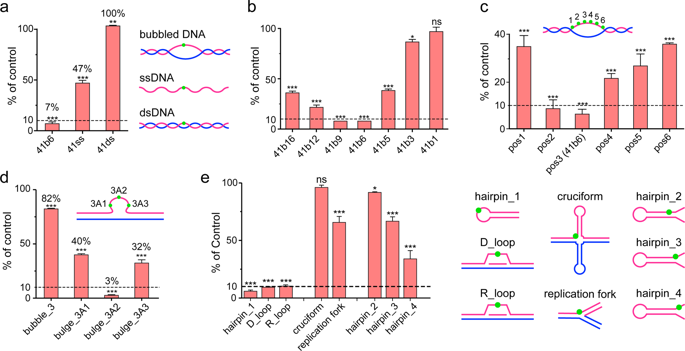
N6-methyladenine (N6-mA) of DNA is an emerging epigenetic mark in mammalian genome. Levels of N6-mA undergo drastic fluctuation during early embryogenesis, indicative of active regulation. Here we show that the 2-oxoglutarate-dependent oxygenase ALKBH1 functions as a nuclear eraser of N6-mA in unpairing regions (e.g., SIDD, Stress-Induced DNA Double Helix Destabilization regions) of mammalian genomes. Enzymatic profiling studies revealed that ALKBH1 prefers bubbled or bulged DNAs as substrate, instead of single-stranded (ss-) or double-stranded (ds-) DNAs. Structural studies of ALKBH1 revealed an unexpected "stretch-out" conformation of its "Flip1" motif, a conserved element that usually bends over catalytic center to facilitate substrate base flipping in other DNA demethylases. Thus, lack of a bending "Flip1" explains the observed preference of ALKBH1 for unpairing substrates, in which the flipped N6-mA is primed for catalysis. Co-crystal structural studies of ALKBH1 bound to a 21-mer bulged DNA explained the need of both flanking duplexes and a flipped base for recognition and catalysis. Key elements (e.g., an ALKBH1-specific α1 helix) as well as residues contributing to structural integrity and catalytic activity were validated by structure-based mutagenesis studies. Furthermore, ssDNA-seq and DIP-seq analyses revealed significant co-occurrence of base unpairing regions with N6-mA in mouse genome. Collectively, our biochemical, structural and genomic studies suggest that ALKBH1 is an important DNA demethylase that regulates genome N6-mA turnover of unpairing regions associated with dynamic chromosome regulation.
中文翻译:

哺乳动物 ALKBH1 用作解配对 DNA 的 N6-mA 去甲基化酶。
DNA 的 N6-甲基腺嘌呤 (N6-mA) 是哺乳动物基因组中新兴的表观遗传标记。N6-mA 的水平在早期胚胎发生过程中发生剧烈波动,表明存在主动调节。在这里,我们表明 2-酮戊二酸依赖性加氧酶 ALKBH1 在哺乳动物基因组的未配对区域(例如,SIDD,应激诱导的 DNA 双螺旋去稳定区域)中充当 N6-mA 的核擦除器。酶促分析研究表明,ALKBH1 更喜欢以气泡或凸起的 DNA 作为底物,而不是单链 (ss-) 或双链 (ds-) DNA。ALKBH1 的结构研究揭示了其“Flip1”基序的意外“伸展”构象,这是一种保守元素,通常在催化中心弯曲以促进其他 DNA 去甲基化酶中的底物碱基翻转。因此,缺少弯曲的“Flip1” 解释了观察到的 ALKBH1 对不配对底物的偏好,其中翻转的 N6-mA 已准备好进行催化。与 21 聚体凸起 DNA 结合的 ALKBH1 的共晶体结构研究解释了侧翼双链体和翻转碱基用于识别和催化的必要性。通过基于结构的诱变研究验证了关键元素(例如,ALKBH1 特异性 α1 螺旋)以及有助于结构完整性和催化活性的残基。此外,ssDNA-seq 和 DIP-seq 分析揭示了小鼠基因组中碱基不配对区域与 N6-mA 的显着共现。总的来说,我们的生化、结构和基因组研究表明,ALKBH1 是一种重要的 DNA 去甲基化酶,可调节与动态染色体调节相关的未配对区域的基因组 N6-mA 转换。
更新日期:2020-02-12
中文翻译:

哺乳动物 ALKBH1 用作解配对 DNA 的 N6-mA 去甲基化酶。
DNA 的 N6-甲基腺嘌呤 (N6-mA) 是哺乳动物基因组中新兴的表观遗传标记。N6-mA 的水平在早期胚胎发生过程中发生剧烈波动,表明存在主动调节。在这里,我们表明 2-酮戊二酸依赖性加氧酶 ALKBH1 在哺乳动物基因组的未配对区域(例如,SIDD,应激诱导的 DNA 双螺旋去稳定区域)中充当 N6-mA 的核擦除器。酶促分析研究表明,ALKBH1 更喜欢以气泡或凸起的 DNA 作为底物,而不是单链 (ss-) 或双链 (ds-) DNA。ALKBH1 的结构研究揭示了其“Flip1”基序的意外“伸展”构象,这是一种保守元素,通常在催化中心弯曲以促进其他 DNA 去甲基化酶中的底物碱基翻转。因此,缺少弯曲的“Flip1” 解释了观察到的 ALKBH1 对不配对底物的偏好,其中翻转的 N6-mA 已准备好进行催化。与 21 聚体凸起 DNA 结合的 ALKBH1 的共晶体结构研究解释了侧翼双链体和翻转碱基用于识别和催化的必要性。通过基于结构的诱变研究验证了关键元素(例如,ALKBH1 特异性 α1 螺旋)以及有助于结构完整性和催化活性的残基。此外,ssDNA-seq 和 DIP-seq 分析揭示了小鼠基因组中碱基不配对区域与 N6-mA 的显着共现。总的来说,我们的生化、结构和基因组研究表明,ALKBH1 是一种重要的 DNA 去甲基化酶,可调节与动态染色体调节相关的未配对区域的基因组 N6-mA 转换。











































 京公网安备 11010802027423号
京公网安备 11010802027423号