当前位置:
X-MOL 学术
›
Nat. Prod. Rep.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Natural allosteric modulators and their biological targets: molecular signatures and mechanisms.
Natural Product Reports ( IF 10.2 ) Pub Date : 2020-02-12 , DOI: 10.1039/c9np00064j Marjorie Bruder 1 , Gina Polo , Daniela B B Trivella
Natural Product Reports ( IF 10.2 ) Pub Date : 2020-02-12 , DOI: 10.1039/c9np00064j Marjorie Bruder 1 , Gina Polo , Daniela B B Trivella
Affiliation
|
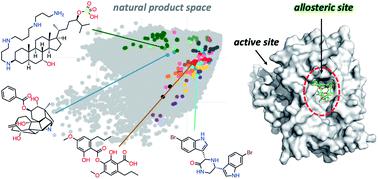
Covering: 2008 to 2018Over the last decade more than two hundred single natural products were confirmed as natural allosteric modulators (alloNPs) of proteins. The compounds are presented and discussed with the support of a chemical space, constructed using a principal component analysis (PCA) of molecular descriptors from chemical compounds of distinct databases. This analysis showed that alloNPs are dispersed throughout the majority of the chemical space defined by natural products in general. Moreover, a cluster of alloNPs was shown to occupy a region almost devoid of allosteric modulators retrieved from a dataset composed mainly of synthetic compounds, further highlighting the importance to explore the entire natural chemical space for probing allosteric mechanisms. The protein targets which alloNPs bind to comprised 81 different proteins, which were classified into 5 major groups, with enzymes, in particular hydrolases, being the main representative group. The review also brings a critical interpretation on the mechanisms by which alloNPs display their molecular action on proteins. In the latter analysis, alloNPs were classified according to their final effect on the target protein, resulting in 3 major categories: (i) local alteration of the orthosteric site; (ii) global alteration in protein dynamics that change function; and (iii) oligomer stabilisation or protein complex destabilisation via protein-protein interaction in sites distant from the orthosteric site. G-protein coupled receptors (GPCRs), which use a combination of the three types of allosteric regulation found, were also probed by natural products. In summary, the natural allosteric modulators reviewed herein emphasise their importance for exploring alternative chemotherapeutic strategies, potentially pushing the boundaries of the druggable space of pharmacologically relevant drug targets.
中文翻译:

天然的变构调节剂及其生物学靶标:分子标记和机理。
研究范围:2008年至2018年在过去十年中,已确认有200多种单一天然产物是蛋白质的天然变构调节剂(alloNPs)。在化学空间的支持下对化合物进行展示和讨论,该化学空间是使用来自不同数据库化合物的分子描述符的主成分分析(PCA)构建的。该分析表明,一般而言,alloNPs分散在天然产物定义的大部分化学空间中。而且,显示出同种异体NP簇占据几乎没有从主要由合成化合物组成的数据集中检索到的变构调节剂的区域,这进一步凸显了探索整个天然化学空间以探测变构机理的重要性。alloNPs结合的蛋白质靶标包含81种不同的蛋白质,它们分为5大类,主要的代表是酶类,尤其是水解酶。该综述还对异源NPs展示其对蛋白质的分子作用的机制提出了重要的解释。在后面的分析中,根据同种异位核苷酸对靶蛋白的最终作用将其分类,可分为3个主要类别:(i)正构位点的局部改变;(ii)改变功能的蛋白质动力学的整体变化;(iii)在远离正构位点的位点通过蛋白质-蛋白质相互作用引起的低聚物稳定或蛋白质复合物不稳定。天然产物也探测了结合了三种变构调节作用的G蛋白偶联受体(GPCR)。综上所述,
更新日期:2020-02-12
中文翻译:

天然的变构调节剂及其生物学靶标:分子标记和机理。
研究范围:2008年至2018年在过去十年中,已确认有200多种单一天然产物是蛋白质的天然变构调节剂(alloNPs)。在化学空间的支持下对化合物进行展示和讨论,该化学空间是使用来自不同数据库化合物的分子描述符的主成分分析(PCA)构建的。该分析表明,一般而言,alloNPs分散在天然产物定义的大部分化学空间中。而且,显示出同种异体NP簇占据几乎没有从主要由合成化合物组成的数据集中检索到的变构调节剂的区域,这进一步凸显了探索整个天然化学空间以探测变构机理的重要性。alloNPs结合的蛋白质靶标包含81种不同的蛋白质,它们分为5大类,主要的代表是酶类,尤其是水解酶。该综述还对异源NPs展示其对蛋白质的分子作用的机制提出了重要的解释。在后面的分析中,根据同种异位核苷酸对靶蛋白的最终作用将其分类,可分为3个主要类别:(i)正构位点的局部改变;(ii)改变功能的蛋白质动力学的整体变化;(iii)在远离正构位点的位点通过蛋白质-蛋白质相互作用引起的低聚物稳定或蛋白质复合物不稳定。天然产物也探测了结合了三种变构调节作用的G蛋白偶联受体(GPCR)。综上所述,











































 京公网安备 11010802027423号
京公网安备 11010802027423号