当前位置:
X-MOL 学术
›
Pharmacogenomics J.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Identification of gene modules associated with survival of diffuse large B-cell lymphoma treated with CHOP-based chemotherapy.
The Pharmacogenomics Journal ( IF 2.9 ) Pub Date : 2020-02-11 , DOI: 10.1038/s41397-020-0161-6 YongChao Gao 1, 2, 3, 4 , Bao Sun 1, 2, 3, 4 , JingLei Hu 1, 2, 3, 4 , Huan Ren 1, 2, 3, 4 , HongHao Zhou 1, 2, 3, 4 , Ling Chen 5 , Rong Liu 1, 2, 3, 4 , Wei Zhang 1, 2, 3, 4
The Pharmacogenomics Journal ( IF 2.9 ) Pub Date : 2020-02-11 , DOI: 10.1038/s41397-020-0161-6 YongChao Gao 1, 2, 3, 4 , Bao Sun 1, 2, 3, 4 , JingLei Hu 1, 2, 3, 4 , Huan Ren 1, 2, 3, 4 , HongHao Zhou 1, 2, 3, 4 , Ling Chen 5 , Rong Liu 1, 2, 3, 4 , Wei Zhang 1, 2, 3, 4
Affiliation
|
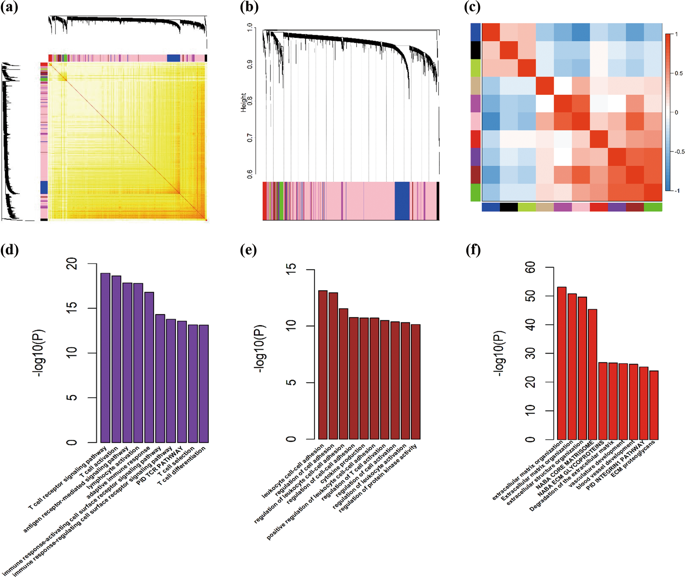
Diffuse Large B-cell Lymphoma (DLBCL), a heterogeneous disease, is influenced by complex network of gene interactions. Most previous studies focused on individual genes, but ignored the importance of intergenic correlations. In current study, we aimed to explore the association between gene networks and overall survival (OS) of DLBCL patients treated with CHOP-based chemotherapy (cyclophosphamide combination with doxorubicin, vincristine and prednisone). Weighted gene co-expression network analysis was conducted to obtain insights into the molecular characteristics of DLBCL. Ten co-expression gene networks (modules) were identified in training dataset (n = 470), and their associations with patients' OS after chemotherapy were tested. The results were validated in four independent datasets (n = 802). Gene ontology (GO) biological function enrichment analysis was conducted with Metascape. Three modules (purple, brown and red), which were enriched in T-cell immune, cell-cell adhesion and extracellular matrix (ECM), respectively, were found to be related to longer OS. Higher expression of several hub genes within these three co-expression modules, for example, LCP2 (HR = 0.77, p = 5.40 × 10-2), CD2 (HR = 0.87, p = 6.31 × 10-2), CD3D (HR = 0.83, p = 6.94 × 10-3), FYB (HR = 0.82, p = 1.40 × 10-2), GZMK (HR = 0.92, p = 1.19 × 10-1), FN1 (HR = 0.88, p = 7.06 × 10-2), SPARC (HR = 0.82, p = 2.06 × 10-2), were found to be associated with favourable survival. Moreover, the associations of the modules and hub genes with OS in different molecular subtypes and different chemotherapy groups were also revealed. In general, our research revealed the key gene modules and several hub genes were upregulated correlated with good survival of DLBCL patients, which might provide potential therapeutic targets for future clinical research.
中文翻译:

鉴定与基于 CHOP 的化疗治疗的弥漫性大 B 细胞淋巴瘤的存活相关的基因模块。
弥漫性大 B 细胞淋巴瘤 (DLBCL) 是一种异质性疾病,受复杂的基因相互作用网络的影响。大多数先前的研究都集中在单个基因上,但忽略了基因间相关性的重要性。在目前的研究中,我们旨在探讨基因网络与接受 CHOP 为基础的化疗(环磷酰胺联合多柔比星、长春新碱和强的松)治疗的 DLBCL 患者的总生存期 (OS) 之间的关联。进行加权基因共表达网络分析以深入了解DLBCL的分子特征。在训练数据集(n = 470)中确定了十个共表达基因网络(模块),并测试了它们与化疗后患者 OS 的关联。结果在四个独立的数据集(n = 802)中得到验证。使用 Metascape 进行基因本体 (GO) 生物功能富集分析。发现分别富含 T 细胞免疫、细胞-细胞粘附和细胞外基质 (ECM) 的三个模块(紫色、棕色和红色)与较长的 OS 相关。在这三个共表达模块中,几个中枢基因的高表达,例如,LCP2 (HR = 0.77, p = 5.40 × 10-2), CD2 (HR = 0.87, p = 6.31 × 10-2), CD3D (HR = 0.83, p = 6.94 × 10-3), FYB (HR = 0.82, p = 1.40 × 10-2), GZMK (HR = 0.92, p = 1.19 × 10-1), FN1 (HR = 0.88, p = 7.06 × 10-2)、SPARC (HR = 0.82, p = 2.06 × 10-2) 被发现与良好的生存率相关。此外,还揭示了不同分子亚型和不同化疗组中模块和枢纽基因与 OS 的关联。一般来说,
更新日期:2020-02-11
中文翻译:

鉴定与基于 CHOP 的化疗治疗的弥漫性大 B 细胞淋巴瘤的存活相关的基因模块。
弥漫性大 B 细胞淋巴瘤 (DLBCL) 是一种异质性疾病,受复杂的基因相互作用网络的影响。大多数先前的研究都集中在单个基因上,但忽略了基因间相关性的重要性。在目前的研究中,我们旨在探讨基因网络与接受 CHOP 为基础的化疗(环磷酰胺联合多柔比星、长春新碱和强的松)治疗的 DLBCL 患者的总生存期 (OS) 之间的关联。进行加权基因共表达网络分析以深入了解DLBCL的分子特征。在训练数据集(n = 470)中确定了十个共表达基因网络(模块),并测试了它们与化疗后患者 OS 的关联。结果在四个独立的数据集(n = 802)中得到验证。使用 Metascape 进行基因本体 (GO) 生物功能富集分析。发现分别富含 T 细胞免疫、细胞-细胞粘附和细胞外基质 (ECM) 的三个模块(紫色、棕色和红色)与较长的 OS 相关。在这三个共表达模块中,几个中枢基因的高表达,例如,LCP2 (HR = 0.77, p = 5.40 × 10-2), CD2 (HR = 0.87, p = 6.31 × 10-2), CD3D (HR = 0.83, p = 6.94 × 10-3), FYB (HR = 0.82, p = 1.40 × 10-2), GZMK (HR = 0.92, p = 1.19 × 10-1), FN1 (HR = 0.88, p = 7.06 × 10-2)、SPARC (HR = 0.82, p = 2.06 × 10-2) 被发现与良好的生存率相关。此外,还揭示了不同分子亚型和不同化疗组中模块和枢纽基因与 OS 的关联。一般来说,











































 京公网安备 11010802027423号
京公网安备 11010802027423号