当前位置:
X-MOL 学术
›
Nat. Microbiol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Rapid inference of antibiotic resistance and susceptibility by genomic neighbour typing.
Nature Microbiology ( IF 20.5 ) Pub Date : 2020-02-10 , DOI: 10.1038/s41564-019-0656-6 Karel Břinda 1, 2 , Alanna Callendrello 1 , Kevin C Ma 3 , Derek R MacFadden 1, 4 , Themoula Charalampous 5 , Robyn S Lee 1, 6 , Lauren Cowley 7 , Crista B Wadsworth 8 , Yonatan H Grad 3 , Gregory Kucherov 9, 10 , Justin O'Grady 5, 11 , Michael Baym 2 , William P Hanage 1
Nature Microbiology ( IF 20.5 ) Pub Date : 2020-02-10 , DOI: 10.1038/s41564-019-0656-6 Karel Břinda 1, 2 , Alanna Callendrello 1 , Kevin C Ma 3 , Derek R MacFadden 1, 4 , Themoula Charalampous 5 , Robyn S Lee 1, 6 , Lauren Cowley 7 , Crista B Wadsworth 8 , Yonatan H Grad 3 , Gregory Kucherov 9, 10 , Justin O'Grady 5, 11 , Michael Baym 2 , William P Hanage 1
Affiliation
|
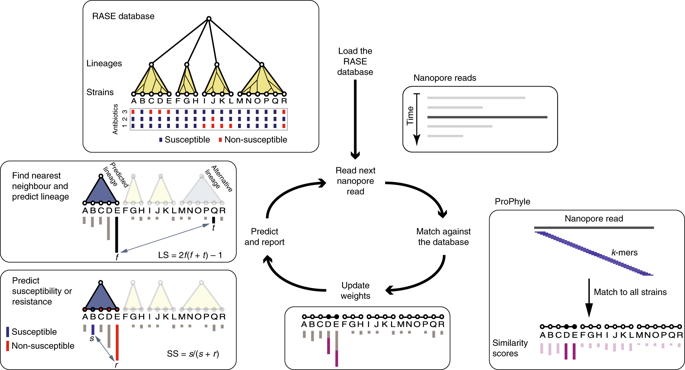
Surveillance of drug-resistant bacteria is essential for healthcare providers to deliver effective empirical antibiotic therapy. However, traditional molecular epidemiology does not typically occur on a timescale that could affect patient treatment and outcomes. Here, we present a method called 'genomic neighbour typing' for inferring the phenotype of a bacterial sample by identifying its closest relatives in a database of genomes with metadata. We show that this technique can infer antibiotic susceptibility and resistance for both Streptococcus pneumoniae and Neisseria gonorrhoeae. We implemented this with rapid k-mer matching, which, when used on Oxford Nanopore MinION data, can run in real time. This resulted in the determination of resistance within 10 min (91% sensitivity and 100% specificity for S. pneumoniae and 81% sensitivity and 100% specificity for N. gonorrhoeae from isolates with a representative database) of starting sequencing, and within 4 h of sample collection (75% sensitivity and 100% specificity for S. pneumoniae) for clinical metagenomic sputum samples. This flexible approach has wide application for pathogen surveillance and may be used to greatly accelerate appropriate empirical antibiotic treatment.
中文翻译:

通过基因组邻居分型快速推断抗生素耐药性和敏感性。
耐药细菌的监测对于医疗保健提供者提供有效的经验性抗生素治疗至关重要。然而,传统的分子流行病学通常不会在可能影响患者治疗和结果的时间范围内发生。在这里,我们提出了一种称为“基因组邻居分型”的方法,通过在带有元数据的基因组数据库中识别其最接近的亲属来推断细菌样本的表型。我们证明,这项技术可以推断肺炎链球菌和淋病奈瑟菌的抗生素敏感性和耐药性。我们通过快速 k-mer 匹配来实现这一点,当将其用于 Oxford Nanopore MinION 数据时,可以实时运行。这导致在开始测序后 10 分钟内(对肺炎链球菌具有 91% 的敏感性和 100% 的特异性,对具有代表性数据库的分离株的淋病奈瑟菌的 81% 的敏感性和 100% 的特异性)内确定了耐药性,并且在开始测序后的 4 小时内确定了耐药性临床宏基因组痰样本的样本采集(肺炎链球菌的敏感性为 75%,特异性为 100%)。这种灵活的方法在病原体监测方面具有广泛的应用,并可用于大大加速适当的经验性抗生素治疗。
更新日期:2020-02-10
中文翻译:

通过基因组邻居分型快速推断抗生素耐药性和敏感性。
耐药细菌的监测对于医疗保健提供者提供有效的经验性抗生素治疗至关重要。然而,传统的分子流行病学通常不会在可能影响患者治疗和结果的时间范围内发生。在这里,我们提出了一种称为“基因组邻居分型”的方法,通过在带有元数据的基因组数据库中识别其最接近的亲属来推断细菌样本的表型。我们证明,这项技术可以推断肺炎链球菌和淋病奈瑟菌的抗生素敏感性和耐药性。我们通过快速 k-mer 匹配来实现这一点,当将其用于 Oxford Nanopore MinION 数据时,可以实时运行。这导致在开始测序后 10 分钟内(对肺炎链球菌具有 91% 的敏感性和 100% 的特异性,对具有代表性数据库的分离株的淋病奈瑟菌的 81% 的敏感性和 100% 的特异性)内确定了耐药性,并且在开始测序后的 4 小时内确定了耐药性临床宏基因组痰样本的样本采集(肺炎链球菌的敏感性为 75%,特异性为 100%)。这种灵活的方法在病原体监测方面具有广泛的应用,并可用于大大加速适当的经验性抗生素治疗。









































 京公网安备 11010802027423号
京公网安备 11010802027423号