当前位置:
X-MOL 学术
›
Genet. Med.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Copy-number variation contributes 9% of pathogenicity in the inherited retinal degenerations.
Genetics in Medicine ( IF 8.8 ) Pub Date : 2020-02-10 , DOI: 10.1038/s41436-020-0759-8 Erin Zampaglione 1 , Benyam Kinde 1 , Emily M Place 1 , Daniel Navarro-Gomez 1 , Matthew Maher 1 , Farzad Jamshidi 1 , Sherwin Nassiri 2 , J Alex Mazzone 1 , Caitlin Finn 1 , Dana Schlegel 3 , Jason Comander 1 , Eric A Pierce 1 , Kinga M Bujakowska 1
Genetics in Medicine ( IF 8.8 ) Pub Date : 2020-02-10 , DOI: 10.1038/s41436-020-0759-8 Erin Zampaglione 1 , Benyam Kinde 1 , Emily M Place 1 , Daniel Navarro-Gomez 1 , Matthew Maher 1 , Farzad Jamshidi 1 , Sherwin Nassiri 2 , J Alex Mazzone 1 , Caitlin Finn 1 , Dana Schlegel 3 , Jason Comander 1 , Eric A Pierce 1 , Kinga M Bujakowska 1
Affiliation
|
PURPOSE
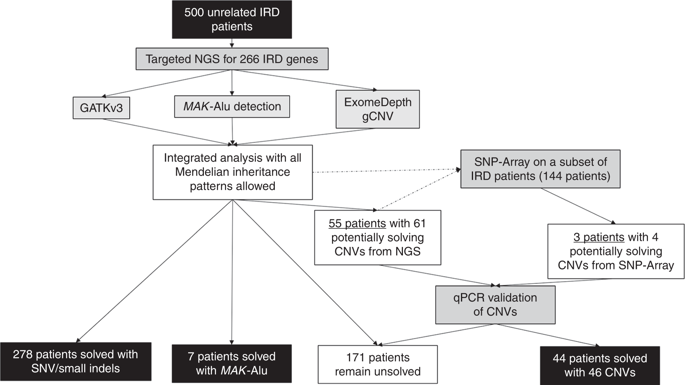
Current sequencing strategies can genetically solve 55-60% of inherited retinal degeneration (IRD) cases, despite recent progress in sequencing. This can partially be attributed to elusive pathogenic variants (PVs) in known IRD genes, including copy-number variations (CNVs), which have been shown as major contributors to unsolved IRD cases.
METHODS
Five hundred IRD patients were analyzed with targeted next-generation sequencing (NGS). The NGS data were used to detect CNVs with ExomeDepth and gCNV and the results were compared with CNV detection with a single-nucleotide polymorphism (SNP) array. Likely causal CNV predictions were validated by quantitative polymerase chain reaction (qPCR).
RESULTS
Likely disease-causing single-nucleotide variants (SNVs) and small indels were found in 55.6% of subjects. PVs in USH2A (11.6%), RPGR (4%), and EYS (4%) were the most common. Likely causal CNVs were found in an additional 8.8% of patients. Of the three CNV detection methods, gCNV showed the highest accuracy. Approximately 30% of unsolved subjects had a single likely PV in a recessive IRD gene.
CONCLUSION
CNV detection using NGS-based algorithms is a reliable method that greatly increases the genetic diagnostic rate of IRDs. Experimentally validating CNVs helps estimate the rate at which IRDs might be solved by a CNV plus a more elusive variant.
中文翻译:

在遗传性视网膜变性中,拷贝数变异占致病性的 9%。
目的 尽管最近在测序方面取得了进展,但目前的测序策略可以从基因上解决 55-60% 的遗传性视网膜变性 (IRD) 病例。这可以部分归因于已知 IRD 基因中难以捉摸的致病变异 (PV),包括拷贝数变异 (CNV),这已被证明是未解决 IRD 病例的主要贡献者。方法 对 500 名 IRD 患者进行了靶向二代测序 (NGS) 分析。NGS 数据用于通过 ExomeDepth 和 gCNV 检测 CNV,并将结果与使用单核苷酸多态性 (SNP) 阵列的 CNV 检测进行比较。通过定量聚合酶链反应 (qPCR) 验证了可能的因果 CNV 预测。结果在 55.6% 的受试者中发现了可能导致疾病的单核苷酸变异 (SNV) 和小插入缺失。USH2A 中的 PV (11.6%)、RPGR (4%)、和 EYS (4%) 是最常见的。在另外 8.8% 的患者中发现了可能的因果 CNV。在三种 CNV 检测方法中,gCNV 的准确度最高。大约 30% 未解决的受试者在隐性 IRD 基因中有一个可能的 PV。结论 使用基于 NGS 的算法检测 CNV 是一种可靠的方法,可大大提高 IRD 的基因诊断率。实验验证 CNV 有助于估计 IRD 可能被 CNV 加上更难以捉摸的变体解决的速率。结论 使用基于 NGS 的算法检测 CNV 是一种可靠的方法,可大大提高 IRD 的基因诊断率。实验验证 CNV 有助于估计 IRD 可能被 CNV 加上更难以捉摸的变体解决的速率。结论 使用基于 NGS 的算法检测 CNV 是一种可靠的方法,可大大提高 IRD 的基因诊断率。实验验证 CNV 有助于估计 IRD 可能被 CNV 加上更难以捉摸的变体解决的速率。
更新日期:2020-02-10
中文翻译:

在遗传性视网膜变性中,拷贝数变异占致病性的 9%。
目的 尽管最近在测序方面取得了进展,但目前的测序策略可以从基因上解决 55-60% 的遗传性视网膜变性 (IRD) 病例。这可以部分归因于已知 IRD 基因中难以捉摸的致病变异 (PV),包括拷贝数变异 (CNV),这已被证明是未解决 IRD 病例的主要贡献者。方法 对 500 名 IRD 患者进行了靶向二代测序 (NGS) 分析。NGS 数据用于通过 ExomeDepth 和 gCNV 检测 CNV,并将结果与使用单核苷酸多态性 (SNP) 阵列的 CNV 检测进行比较。通过定量聚合酶链反应 (qPCR) 验证了可能的因果 CNV 预测。结果在 55.6% 的受试者中发现了可能导致疾病的单核苷酸变异 (SNV) 和小插入缺失。USH2A 中的 PV (11.6%)、RPGR (4%)、和 EYS (4%) 是最常见的。在另外 8.8% 的患者中发现了可能的因果 CNV。在三种 CNV 检测方法中,gCNV 的准确度最高。大约 30% 未解决的受试者在隐性 IRD 基因中有一个可能的 PV。结论 使用基于 NGS 的算法检测 CNV 是一种可靠的方法,可大大提高 IRD 的基因诊断率。实验验证 CNV 有助于估计 IRD 可能被 CNV 加上更难以捉摸的变体解决的速率。结论 使用基于 NGS 的算法检测 CNV 是一种可靠的方法,可大大提高 IRD 的基因诊断率。实验验证 CNV 有助于估计 IRD 可能被 CNV 加上更难以捉摸的变体解决的速率。结论 使用基于 NGS 的算法检测 CNV 是一种可靠的方法,可大大提高 IRD 的基因诊断率。实验验证 CNV 有助于估计 IRD 可能被 CNV 加上更难以捉摸的变体解决的速率。



























 京公网安备 11010802027423号
京公网安备 11010802027423号