当前位置:
X-MOL 学术
›
ACS Synth. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Mathematical Models of Protease-Based Enzymatic Biosensors.
ACS Synthetic Biology ( IF 3.7 ) Pub Date : 2020-02-07 , DOI: 10.1021/acssynbio.9b00279 Deepak K Agrawal 1, 2 , Elliott M Dolan 3 , Nancy E Hernandez 3 , Kristin M Blacklock 3 , Sagar D Khare 3 , Eduardo D Sontag 1, 2, 4
ACS Synthetic Biology ( IF 3.7 ) Pub Date : 2020-02-07 , DOI: 10.1021/acssynbio.9b00279 Deepak K Agrawal 1, 2 , Elliott M Dolan 3 , Nancy E Hernandez 3 , Kristin M Blacklock 3 , Sagar D Khare 3 , Eduardo D Sontag 1, 2, 4
Affiliation
|
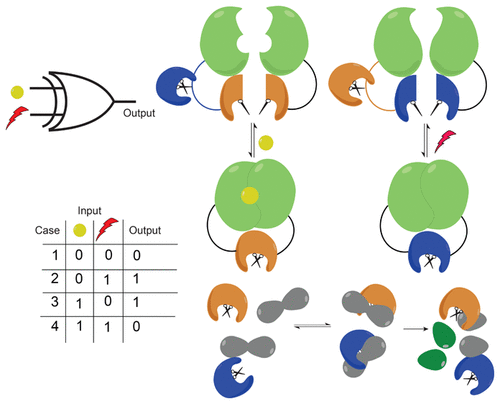
An important goal of synthetic biology is to build biosensors and circuits with well-defined input-output relationships that operate at speeds found in natural biological systems. However, for molecular computation, most commonly used genetic circuit elements typically involve several steps from input detection to output signal production: transcription, translation, and post-translational modifications. These multiple steps together require up to several hours to respond to a single stimulus, and this limits the overall speed and complexity of genetic circuits. To address this gap, molecular frameworks that rely exclusively on post-translational steps to realize reaction networks that can process inputs at a time scale of seconds to minutes have been proposed. Here, we build mathematical models of fast biosensors capable of producing Boolean logic functionality. We employ protease-based chemical and light-induced switches, investigate their operation, and provide selection guidelines for their use as on-off switches. As a proof of concept, we implement a rapamycin-induced switch in vitro and demonstrate that its response qualitatively agrees with the predictions from our models. We then use these switches as elementary blocks, developing models for biosensors that can perform OR and XOR Boolean logic computation while using reaction conditions as tuning parameters. We use sensitivity analysis to determine the time-dependent sensitivity of the output to proteolytic and protein-protein binding reaction parameters. These fast protease-based biosensors can be used to implement complex molecular circuits with a capability of processing multiple inputs controllably and algorithmically. Our framework for evaluating and optimizing circuit performance can be applied to other molecular logic circuits.
中文翻译:

基于蛋白酶的酶生物传感器的数学模型。
合成生物学的一个重要目标是构建具有明确定义的输入-输出关系的生物传感器和电路,并以自然生物系统中发现的速度运行。但是,对于分子计算,最常用的遗传电路元件通常涉及从输入检测到输出信号产生的几个步骤:转录,翻译和翻译后修饰。这些多个步骤一起需要多达几个小时才能对单个刺激做出响应,这限制了遗传回路的整体速度和复杂性。为了解决这个空白,已经提出了仅依赖于翻译后步骤来实现可以在数秒至数分钟的时间范围内处理输入的反应网络的分子框架。这里,我们建立了能够产生布尔逻辑功能的快速生物传感器的数学模型。我们使用基于蛋白酶的化学和光诱导开关,研究它们的操作,并提供选择指南作为它们的开关使用。作为概念的证明,我们在体外实施雷帕霉素诱导的开关,并证明其反应在质量上与我们的模型预测相符。然后,我们将这些开关用作基本模块,为生物传感器开发模型,该模型可以在使用反应条件作为调整参数的同时执行OR和XOR布尔逻辑计算。我们使用敏感性分析来确定输出对蛋白水解和蛋白质-蛋白质结合反应参数的时间依赖性敏感性。这些基于蛋白酶的快速生物传感器可用于实现复杂的分子电路,并具有可控地和算法地处理多个输入的能力。我们用于评估和优化电路性能的框架可以应用于其他分子逻辑电路。
更新日期:2020-02-07
中文翻译:

基于蛋白酶的酶生物传感器的数学模型。
合成生物学的一个重要目标是构建具有明确定义的输入-输出关系的生物传感器和电路,并以自然生物系统中发现的速度运行。但是,对于分子计算,最常用的遗传电路元件通常涉及从输入检测到输出信号产生的几个步骤:转录,翻译和翻译后修饰。这些多个步骤一起需要多达几个小时才能对单个刺激做出响应,这限制了遗传回路的整体速度和复杂性。为了解决这个空白,已经提出了仅依赖于翻译后步骤来实现可以在数秒至数分钟的时间范围内处理输入的反应网络的分子框架。这里,我们建立了能够产生布尔逻辑功能的快速生物传感器的数学模型。我们使用基于蛋白酶的化学和光诱导开关,研究它们的操作,并提供选择指南作为它们的开关使用。作为概念的证明,我们在体外实施雷帕霉素诱导的开关,并证明其反应在质量上与我们的模型预测相符。然后,我们将这些开关用作基本模块,为生物传感器开发模型,该模型可以在使用反应条件作为调整参数的同时执行OR和XOR布尔逻辑计算。我们使用敏感性分析来确定输出对蛋白水解和蛋白质-蛋白质结合反应参数的时间依赖性敏感性。这些基于蛋白酶的快速生物传感器可用于实现复杂的分子电路,并具有可控地和算法地处理多个输入的能力。我们用于评估和优化电路性能的框架可以应用于其他分子逻辑电路。











































 京公网安备 11010802027423号
京公网安备 11010802027423号