当前位置:
X-MOL 学术
›
Genet. Med.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Noninvasive prenatal diagnosis by genome-wide haplotyping of cell-free plasma DNA.
Genetics in Medicine ( IF 6.6 ) Pub Date : 2020-02-06 , DOI: 10.1038/s41436-019-0748-y Huiwen Che 1 , Darine Villela 1, 2 , Eftychia Dimitriadou 1 , Cindy Melotte 1 , Nathalie Brison 1 , Maria Neofytou 1 , Kris Van Den Bogaert 1 , Olga Tsuiko 1 , Koen Devriendt 1 , Eric Legius 1 , Masoud Zamani Esteki 1, 3, 4 , Thierry Voet 1 , Joris Robert Vermeesch 1
Genetics in Medicine ( IF 6.6 ) Pub Date : 2020-02-06 , DOI: 10.1038/s41436-019-0748-y Huiwen Che 1 , Darine Villela 1, 2 , Eftychia Dimitriadou 1 , Cindy Melotte 1 , Nathalie Brison 1 , Maria Neofytou 1 , Kris Van Den Bogaert 1 , Olga Tsuiko 1 , Koen Devriendt 1 , Eric Legius 1 , Masoud Zamani Esteki 1, 3, 4 , Thierry Voet 1 , Joris Robert Vermeesch 1
Affiliation
|
PURPOSE
Whereas noninvasive prenatal screening for aneuploidies is widely implemented, there is an increasing need for universal approaches for noninvasive prenatal screening for monogenic diseases. Here, we present a cost-effective, generic cell-free fetal DNA (cffDNA) haplotyping approach to scan the fetal genome for the presence of inherited monogenic diseases.
METHODS
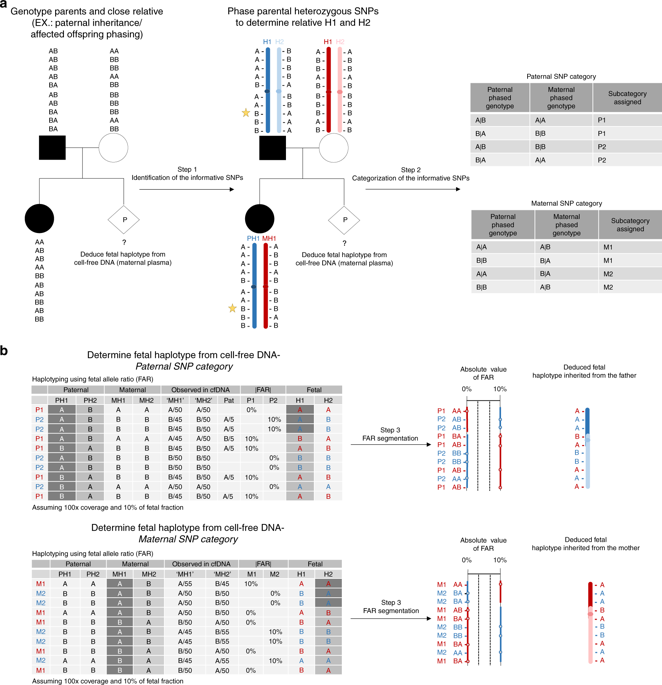
Families participating in the preimplantation genetic testing for monogenic disorders (PGT-M) program were recruited for this study. Two hundred fifty thousand single-nucleotide polymorphisms (SNPs) captured from maternal plasma DNA along with genomic DNA from family members were massively parallel sequenced. Parental genotypes were phased via an available genotype from a close relative, and the fetal genome-wide haplotype and copy number were determined using cffDNA haplotyping analysis based on estimation and segmentation of fetal allele presence in the maternal plasma.
RESULTS
In all families tested, mutational profiles from cffDNA haplotyping are consistent with embryo biopsy profiles. Genome-wide fetal haplotypes are on average 97% concordant with the newborn haplotypes and embryo haplotypes.
CONCLUSION
We demonstrate that genome-wide targeted capture and sequencing of polymorphic SNPs from maternal plasma cell-free DNA (cfDNA) allows haplotyping and copy-number profiling of the fetal genome during pregnancy. The method enables the accurate reconstruction of the fetal haplotypes and can be easily implemented in clinical practice.
中文翻译:

通过无细胞血浆 DNA 的全基因组单倍型进行无创产前诊断。
目的 尽管非整倍体的无创产前筛查已广泛实施,但对单基因疾病无创产前筛查的通用方法的需求日益增加。在这里,我们提出了一种具有成本效益的通用无细胞胎儿 DNA (cffDNA) 单倍型方法来扫描胎儿基因组中是否存在遗传性单基因疾病。方法 参与单基因疾病植入前基因检测 (PGT-M) 计划的家庭被招募参与这项研究。对从母体血浆 DNA 中捕获的 25 万个单核苷酸多态性 (SNP) 以及来自家族成员的基因组 DNA 进行大规模平行测序。亲本基因型是通过近亲的可用基因型分阶段的,基于母体血浆中胎儿等位基因存在的估计和分割,使用 cffDNA 单倍型分析确定胎儿全基因组单倍型和拷贝数。结果 在所有测试的家庭中,来自 cffDNA 单倍型的突变谱与胚胎活检谱一致。全基因组胎儿单倍型与新生儿单倍型和胚胎单倍型的一致性平均为 97%。结论 我们证明,全基因组靶向捕获和测序来自母体无浆细胞 DNA (cfDNA) 的多态性 SNP 可以在怀孕期间对胎儿基因组进行单倍型分析和拷贝数分析。该方法能够准确地重建胎儿单倍型,并且可以在临床实践中容易地实施。结果 在所有测试的家庭中,来自 cffDNA 单倍型的突变谱与胚胎活检谱一致。全基因组胎儿单倍型与新生儿单倍型和胚胎单倍型的一致性平均为 97%。结论 我们证明,全基因组靶向捕获和测序来自母体无浆细胞 DNA (cfDNA) 的多态性 SNP 可以在怀孕期间对胎儿基因组进行单倍型分析和拷贝数分析。该方法能够准确地重建胎儿单倍型,并且可以在临床实践中容易地实施。结果 在所有测试的家庭中,来自 cffDNA 单倍型的突变谱与胚胎活检谱一致。全基因组胎儿单倍型与新生儿单倍型和胚胎单倍型的一致性平均为 97%。结论 我们证明,全基因组靶向捕获和测序来自母体无浆细胞 DNA (cfDNA) 的多态性 SNP 可以在怀孕期间对胎儿基因组进行单倍型分析和拷贝数分析。该方法能够准确地重建胎儿单倍型,并且可以在临床实践中容易地实施。结论 我们证明,全基因组靶向捕获和测序来自母体无浆细胞 DNA (cfDNA) 的多态性 SNP 可以在怀孕期间对胎儿基因组进行单倍型分析和拷贝数分析。该方法能够准确地重建胎儿单倍型,并且可以在临床实践中容易地实施。结论 我们证明,全基因组靶向捕获和测序来自母体无浆细胞 DNA (cfDNA) 的多态性 SNP 可以在怀孕期间对胎儿基因组进行单倍型分析和拷贝数分析。该方法能够准确地重建胎儿单倍型,并且可以在临床实践中容易地实施。
更新日期:2020-02-06
中文翻译:

通过无细胞血浆 DNA 的全基因组单倍型进行无创产前诊断。
目的 尽管非整倍体的无创产前筛查已广泛实施,但对单基因疾病无创产前筛查的通用方法的需求日益增加。在这里,我们提出了一种具有成本效益的通用无细胞胎儿 DNA (cffDNA) 单倍型方法来扫描胎儿基因组中是否存在遗传性单基因疾病。方法 参与单基因疾病植入前基因检测 (PGT-M) 计划的家庭被招募参与这项研究。对从母体血浆 DNA 中捕获的 25 万个单核苷酸多态性 (SNP) 以及来自家族成员的基因组 DNA 进行大规模平行测序。亲本基因型是通过近亲的可用基因型分阶段的,基于母体血浆中胎儿等位基因存在的估计和分割,使用 cffDNA 单倍型分析确定胎儿全基因组单倍型和拷贝数。结果 在所有测试的家庭中,来自 cffDNA 单倍型的突变谱与胚胎活检谱一致。全基因组胎儿单倍型与新生儿单倍型和胚胎单倍型的一致性平均为 97%。结论 我们证明,全基因组靶向捕获和测序来自母体无浆细胞 DNA (cfDNA) 的多态性 SNP 可以在怀孕期间对胎儿基因组进行单倍型分析和拷贝数分析。该方法能够准确地重建胎儿单倍型,并且可以在临床实践中容易地实施。结果 在所有测试的家庭中,来自 cffDNA 单倍型的突变谱与胚胎活检谱一致。全基因组胎儿单倍型与新生儿单倍型和胚胎单倍型的一致性平均为 97%。结论 我们证明,全基因组靶向捕获和测序来自母体无浆细胞 DNA (cfDNA) 的多态性 SNP 可以在怀孕期间对胎儿基因组进行单倍型分析和拷贝数分析。该方法能够准确地重建胎儿单倍型,并且可以在临床实践中容易地实施。结果 在所有测试的家庭中,来自 cffDNA 单倍型的突变谱与胚胎活检谱一致。全基因组胎儿单倍型与新生儿单倍型和胚胎单倍型的一致性平均为 97%。结论 我们证明,全基因组靶向捕获和测序来自母体无浆细胞 DNA (cfDNA) 的多态性 SNP 可以在怀孕期间对胎儿基因组进行单倍型分析和拷贝数分析。该方法能够准确地重建胎儿单倍型,并且可以在临床实践中容易地实施。结论 我们证明,全基因组靶向捕获和测序来自母体无浆细胞 DNA (cfDNA) 的多态性 SNP 可以在怀孕期间对胎儿基因组进行单倍型分析和拷贝数分析。该方法能够准确地重建胎儿单倍型,并且可以在临床实践中容易地实施。结论 我们证明,全基因组靶向捕获和测序来自母体无浆细胞 DNA (cfDNA) 的多态性 SNP 可以在怀孕期间对胎儿基因组进行单倍型分析和拷贝数分析。该方法能够准确地重建胎儿单倍型,并且可以在临床实践中容易地实施。











































 京公网安备 11010802027423号
京公网安备 11010802027423号