Computational and Structural Biotechnology Journal ( IF 4.4 ) Pub Date : 2019-11-30 , DOI: 10.1016/j.csbj.2019.11.004 Hai-Cheng Yi 1, 2 , Zhu-Hong You 1 , Li Cheng 1 , Xi Zhou 1 , Tong-Hai Jiang 1 , Xiao Li 1 , Yan-Bin Wang 1
|
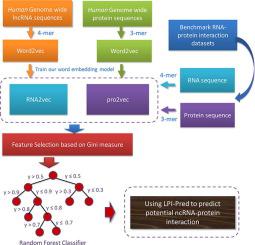
The long noncoding RNAs (lncRNAs) are ubiquitous in organisms and play crucial role in a variety of biological processes and complex diseases. Emerging evidences suggest that lncRNAs interact with corresponding proteins to perform their regulatory functions. Therefore, identifying interacting lncRNA-protein pairs is the first step in understanding the function and mechanism of lncRNA. Since it is time-consuming and expensive to determine lncRNA-protein interactions by high-throughput experiments, more robust and accurate computational methods need to be developed. In this study, we developed a new sequence distributed representation learning based method for potential lncRNA-Protein Interactions Prediction, named LPI-Pred, which is inspired by the similarity between natural language and biological sequences. More specifically, lncRNA and protein sequences were divided into k-mer segmentation, which can be regard as “word” in natural language processing. Then, we trained out the RNA2vec and Pro2vec model using word2vec and human genome-wide lncRNA and protein sequences to mine distribution representation of RNA and protein. Then, the dimension of complex features is reduced by using feature selection based on Gini information impurity measure. Finally, these discriminative features are used to train a Random Forest classifier to predict lncRNA-protein interactions. Five-fold cross-validation was adopted to evaluate the performance of LPI-Pred on three benchmark datasets, including RPI369, RPI488 and RPI2241. The results demonstrate that LPI-Pred can be a useful tool to provide reliable guidance for biological research.
中文翻译:

学习RNA和蛋白质序列的分布式表示及其在预测lncRNA-蛋白质相互作用中的应用
长的非编码RNA(lncRNA)在生物体中无处不在,并在各种生物过程和复杂疾病中发挥关键作用。新兴证据表明,lncRNA与相应的蛋白质相互作用以执行其调节功能。因此,鉴定相互作用的lncRNA-蛋白质对是理解lncRNA的功能和机制的第一步。由于通过高通量实验确定lncRNA-蛋白质相互作用既耗时又昂贵,因此需要开发更强大和准确的计算方法。在这项研究中,我们开发了一种新的基于序列分布式表示学习的潜在lncRNA-蛋白质相互作用预测的方法,称为LPI-Pred,其灵感来自自然语言和生物学序列之间的相似性。进一步来说,k- mer分割,在自然语言处理中可以被视为“单词”。然后,我们使用word2vec和人类全基因组lncRNA和蛋白质序列训练出RNA2vec和Pro2vec模型,以挖掘RNA和蛋白质的分布表示形式。然后,通过使用基于基尼信息杂质测度的特征选择来减小复杂特征的维数。最后,这些判别特征用于训练随机森林分类器以预测lncRNA-蛋白质相互作用。采用五重交叉验证在三个基准数据集(包括RPI369,RPI488和RPI2241)上评估LPI-Pred的性能。结果表明,LPI-Pred可以作为有用的工具,为生物学研究提供可靠的指导。











































 京公网安备 11010802027423号
京公网安备 11010802027423号