当前位置:
X-MOL 学术
›
Pharmacogenomics J.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Effect of genetic polymorphisms in CYP3A4, CYP3A5, and m-TOR on everolimus blood exposure and clinical outcomes in cancer patients.
The Pharmacogenomics Journal ( IF 2.9 ) Pub Date : 2020-02-04 , DOI: 10.1038/s41397-020-0152-7 Stéphanie Bonnet 1, 2 , Sabrina Falkowski 3 , Marine Deppenweiler 4 , Caroline Monchaud 1, 2, 5 , Hélène Arnion 1, 2 , Nicolas Picard 1, 2, 5 , Jean-Baptiste Woillard 1, 2, 5
The Pharmacogenomics Journal ( IF 2.9 ) Pub Date : 2020-02-04 , DOI: 10.1038/s41397-020-0152-7 Stéphanie Bonnet 1, 2 , Sabrina Falkowski 3 , Marine Deppenweiler 4 , Caroline Monchaud 1, 2, 5 , Hélène Arnion 1, 2 , Nicolas Picard 1, 2, 5 , Jean-Baptiste Woillard 1, 2, 5
Affiliation
|
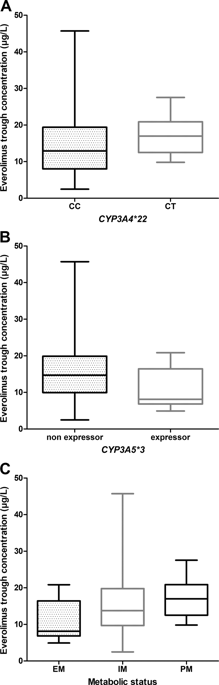
Genetic variations in CYP3A4, CYP3A5, and m-TOR could contribute to interpatient variability regarding m-TOR inhibitors pharmacokinetics or cellular effects. The purpose of this study was to evaluate the influence of selected candidate variations in these genes on everolimus pharmacokinetics, efficacy, and toxicity in cancer patients. Thirty-four patients receiving everolimus for breast (n = 22) or renal (n = 10) cancers, or neuroendocrine tumors of pancreatic origin (n = 2) were included in the study. Six variants in genes related to everolimus pharmacokinetics (CYP3A4*22 and CYP3A5*3) or pharmacodynamics (m-TOR rs2295079, rs2295080, rs2024627 and rs1057079) were genotyped. Associations with trough concentrations (C0), dose reductions, or treatment interruptions due to toxicity and progression-free survival were investigated using generalized estimating equations and Cox models. CYP3A5 nonexpressers had significantly higher C0 as compared with expressers (βGG vs AG = + 6.32 ± 2.22 ng/mL, p = 0.004). m-TOR rs2024627 was significantly associated with an increased risk of cancer progression studied alone or as part of an haplotype (T vs C: HR = 2.60, 95% CI [1.16-5.80], p = 0.020; CTCG vs other haplotypes HR = 2.29, 95% CI [1.06-4.95], p = 0.035, respectively). This study showed that CYP3A5 expression impacts everolimus pharmacokinetics in cancer patients and identified a genetic variation in m-TOR associated with the risk of cancer progression.
中文翻译:

CYP3A4,CYP3A5和m-TOR基因多态性对依维莫司血液暴露和癌症患者临床结局的影响。
CYP3A4,CYP3A5和m-TOR的遗传变异可能会导致患者间关于m-TOR抑制剂药代动力学或细胞作用的变异性。这项研究的目的是评估这些基因中选定的候选变异对依维莫司药代动力学,疗效和对癌症患者的毒性的影响。该研究包括了34名接受依维莫司治疗的乳腺癌(n = 22)或肾癌(n = 10)或胰腺源性神经内分泌肿瘤(n = 2)患者。对依维莫司药代动力学(CYP3A4 * 22和CYP3A5 * 3)或药代动力学(m-TOR rs2295079,rs2295080,rs2024627和rs1057079)相关基因的六个变体进行基因分型。与谷浓度(C0),剂量减少,使用广义估计方程和Cox模型研究由于毒性和无进展生存而导致的治疗中断或治疗中断。CYP3A5非表达者的CO明显高于表达者(βGGvs AG = + 6.32±2.22 ng / mL,p = 0.004)。m-TOR rs2024627与单独或作为单倍型研究的癌症进展风险增加显着相关(T vs C:HR = 2.60,95%CI [1.16-5.80],p = 0.020; CTCG vs其他单倍型HR = 2.29,95%CI [1.06-4.95],p = 0.035)。这项研究表明CYP3A5表达影响癌症患者的依维莫司药代动力学,并确定了m-TOR的遗传变异与癌症进展的风险有关。22 ng / mL,p = 0.004)。m-TOR rs2024627与单独或作为单倍型研究的癌症进展风险增加显着相关(T vs C:HR = 2.60,95%CI [1.16-5.80],p = 0.020; CTCG vs其他单倍型HR = 2.29,95%CI [1.06-4.95],p = 0.035)。这项研究表明CYP3A5表达影响癌症患者的依维莫司药代动力学,并确定了m-TOR的遗传变异与癌症进展的风险有关。22 ng / mL,p = 0.004)。m-TOR rs2024627与单独或作为单倍型研究的癌症进展风险增加显着相关(T vs C:HR = 2.60,95%CI [1.16-5.80],p = 0.020; CTCG vs其他单倍型HR = 2.29,95%CI [1.06-4.95],p = 0.035)。这项研究表明CYP3A5表达影响癌症患者的依维莫司药代动力学,并确定了m-TOR的遗传变异与癌症进展的风险有关。
更新日期:2020-02-04
中文翻译:

CYP3A4,CYP3A5和m-TOR基因多态性对依维莫司血液暴露和癌症患者临床结局的影响。
CYP3A4,CYP3A5和m-TOR的遗传变异可能会导致患者间关于m-TOR抑制剂药代动力学或细胞作用的变异性。这项研究的目的是评估这些基因中选定的候选变异对依维莫司药代动力学,疗效和对癌症患者的毒性的影响。该研究包括了34名接受依维莫司治疗的乳腺癌(n = 22)或肾癌(n = 10)或胰腺源性神经内分泌肿瘤(n = 2)患者。对依维莫司药代动力学(CYP3A4 * 22和CYP3A5 * 3)或药代动力学(m-TOR rs2295079,rs2295080,rs2024627和rs1057079)相关基因的六个变体进行基因分型。与谷浓度(C0),剂量减少,使用广义估计方程和Cox模型研究由于毒性和无进展生存而导致的治疗中断或治疗中断。CYP3A5非表达者的CO明显高于表达者(βGGvs AG = + 6.32±2.22 ng / mL,p = 0.004)。m-TOR rs2024627与单独或作为单倍型研究的癌症进展风险增加显着相关(T vs C:HR = 2.60,95%CI [1.16-5.80],p = 0.020; CTCG vs其他单倍型HR = 2.29,95%CI [1.06-4.95],p = 0.035)。这项研究表明CYP3A5表达影响癌症患者的依维莫司药代动力学,并确定了m-TOR的遗传变异与癌症进展的风险有关。22 ng / mL,p = 0.004)。m-TOR rs2024627与单独或作为单倍型研究的癌症进展风险增加显着相关(T vs C:HR = 2.60,95%CI [1.16-5.80],p = 0.020; CTCG vs其他单倍型HR = 2.29,95%CI [1.06-4.95],p = 0.035)。这项研究表明CYP3A5表达影响癌症患者的依维莫司药代动力学,并确定了m-TOR的遗传变异与癌症进展的风险有关。22 ng / mL,p = 0.004)。m-TOR rs2024627与单独或作为单倍型研究的癌症进展风险增加显着相关(T vs C:HR = 2.60,95%CI [1.16-5.80],p = 0.020; CTCG vs其他单倍型HR = 2.29,95%CI [1.06-4.95],p = 0.035)。这项研究表明CYP3A5表达影响癌症患者的依维莫司药代动力学,并确定了m-TOR的遗传变异与癌症进展的风险有关。











































 京公网安备 11010802027423号
京公网安备 11010802027423号