当前位置:
X-MOL 学术
›
Transl. Psychiaty
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
A polygenic predictor of treatment-resistant depression using whole exome sequencing and genome-wide genotyping.
Translational Psychiatry ( IF 5.8 ) Pub Date : 2020-02-03 , DOI: 10.1038/s41398-020-0738-5 Chiara Fabbri 1 , Siegfried Kasper 2 , Alexander Kautzky 2 , Joseph Zohar 3 , Daniel Souery 4 , Stuart Montgomery 5 , Diego Albani 6 , Gianluigi Forloni 6 , Panagiotis Ferentinos 7 , Dan Rujescu 8 , Julien Mendlewicz 9 , Rudolf Uher 10 , Cathryn M Lewis 1 , Alessandro Serretti 11
Translational Psychiatry ( IF 5.8 ) Pub Date : 2020-02-03 , DOI: 10.1038/s41398-020-0738-5 Chiara Fabbri 1 , Siegfried Kasper 2 , Alexander Kautzky 2 , Joseph Zohar 3 , Daniel Souery 4 , Stuart Montgomery 5 , Diego Albani 6 , Gianluigi Forloni 6 , Panagiotis Ferentinos 7 , Dan Rujescu 8 , Julien Mendlewicz 9 , Rudolf Uher 10 , Cathryn M Lewis 1 , Alessandro Serretti 11
Affiliation
|
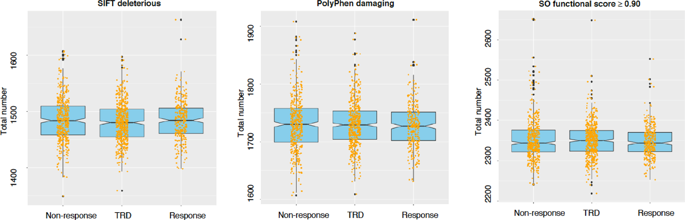
Treatment-resistant depression (TRD) occurs in ~30% of patients with major depressive disorder (MDD) but the genetics of TRD was previously poorly investigated. Whole exome sequencing and genome-wide genotyping were available in 1209 MDD patients after quality control. Antidepressant response was compared to non-response to one treatment and non-response to two or more treatments (TRD). Differences in the risk of carrying damaging variants were tested. A score expressing the burden of variants in genes and pathways was calculated weighting each variant for its functional (Eigen) score and frequency. Gene-based and pathway-based scores were used to develop predictive models of TRD and non-response using gradient boosting in 70% of the sample (training) which were tested in the remaining 30% (testing), evaluating also the addition of clinical predictors. Independent replication was tested in STAR*D and GENDEP using exome array-based data. TRD and non-responders did not show higher risk to carry damaging variants compared to responders. Genes/pathways associated with TRD included those modulating cell survival and proliferation, neurodegeneration, and immune response. Genetic models showed significant prediction of TRD vs. response and they were improved by the addition of clinical predictors, but they were not significantly better than clinical predictors alone. Replication results were driven by clinical factors, except for a model developed in subjects treated with serotonergic antidepressants, which showed a clear improvement in prediction at the extremes of the genetic score distribution in STAR*D. These results suggested relevant biological mechanisms implicated in TRD and a new methodological approach to the prediction of TRD.
中文翻译:

使用整个外显子组测序和全基因组基因分型的多药抗药性抑郁症预测因子。
约30%的重度抑郁症(MDD)患者发生抗药性抑郁症(TRD),但以前对TRD的遗传学研究不足。经过质量控制后,在1209名MDD患者中可获得完整的外显子组测序和全基因组基因分型。将抗抑郁药的反应与对一种治疗的无反应和对两种或多种治疗的无反应(TRD)进行比较。测试了携带破坏性变异体的风险差异。计算表达基因和途径中变体负担的分数,对每个变体的功能(本征)分数和频率加权。使用基于基因和基于路径的评分,在70%的样本(训练)中使用梯度增强来建立TRD和无应答的预测模型,其余30%(测试)中进行了测试,还评估其他临床预测指标。使用基于外显子组阵列的数据在STAR * D和GENDEP中测试了独立复制。与响应者相比,TRD和非响应者携带破坏性变体的风险更高。与TRD相关的基因/途径包括调节细胞存活和增殖,神经变性和免疫反应的基因/途径。遗传模型显示出TRD与响应之间的显着预测,并且通过添加临床预测因子可以改善遗传模型,但它们并不比单独的临床预测因子显着更好。复制结果是受临床因素驱动的,除了在接受血清素能抗抑郁药治疗的受试者中开发的模型外,该模型在STAR * D基因评分分布的极端情况下显示出明显的预测改善。
更新日期:2020-02-03
中文翻译:

使用整个外显子组测序和全基因组基因分型的多药抗药性抑郁症预测因子。
约30%的重度抑郁症(MDD)患者发生抗药性抑郁症(TRD),但以前对TRD的遗传学研究不足。经过质量控制后,在1209名MDD患者中可获得完整的外显子组测序和全基因组基因分型。将抗抑郁药的反应与对一种治疗的无反应和对两种或多种治疗的无反应(TRD)进行比较。测试了携带破坏性变异体的风险差异。计算表达基因和途径中变体负担的分数,对每个变体的功能(本征)分数和频率加权。使用基于基因和基于路径的评分,在70%的样本(训练)中使用梯度增强来建立TRD和无应答的预测模型,其余30%(测试)中进行了测试,还评估其他临床预测指标。使用基于外显子组阵列的数据在STAR * D和GENDEP中测试了独立复制。与响应者相比,TRD和非响应者携带破坏性变体的风险更高。与TRD相关的基因/途径包括调节细胞存活和增殖,神经变性和免疫反应的基因/途径。遗传模型显示出TRD与响应之间的显着预测,并且通过添加临床预测因子可以改善遗传模型,但它们并不比单独的临床预测因子显着更好。复制结果是受临床因素驱动的,除了在接受血清素能抗抑郁药治疗的受试者中开发的模型外,该模型在STAR * D基因评分分布的极端情况下显示出明显的预测改善。











































 京公网安备 11010802027423号
京公网安备 11010802027423号