当前位置:
X-MOL 学术
›
Nat. Microbiol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Spatial metabolomics of in situ host-microbe interactions at the micrometre scale.
Nature Microbiology ( IF 28.3 ) Pub Date : 2020-02-03 , DOI: 10.1038/s41564-019-0664-6 Benedikt Geier 1 , Emilia M Sogin 1 , Dolma Michellod 1 , Moritz Janda 1 , Mario Kompauer 2 , Bernhard Spengler 2 , Nicole Dubilier 1, 3 , Manuel Liebeke 1
Nature Microbiology ( IF 28.3 ) Pub Date : 2020-02-03 , DOI: 10.1038/s41564-019-0664-6 Benedikt Geier 1 , Emilia M Sogin 1 , Dolma Michellod 1 , Moritz Janda 1 , Mario Kompauer 2 , Bernhard Spengler 2 , Nicole Dubilier 1, 3 , Manuel Liebeke 1
Affiliation
|
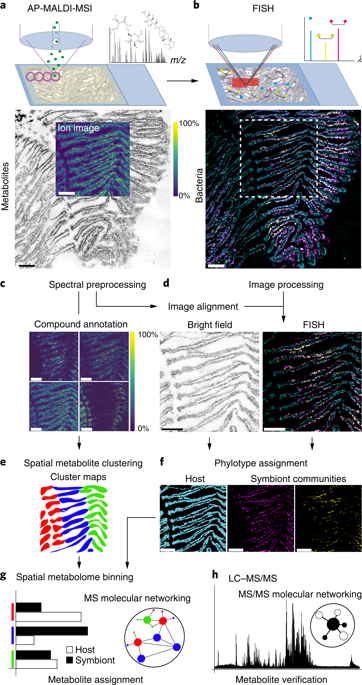
Spatial metabolomics describes the location and chemistry of small molecules involved in metabolic phenotypes, defence molecules and chemical interactions in natural communities. Most current techniques are unable to spatially link the genotype and metabolic phenotype of microorganisms in situ at a scale relevant to microbial interactions. Here, we present a spatial metabolomics pipeline (metaFISH) that combines fluorescence in situ hybridization (FISH) microscopy and high-resolution atmospheric-pressure matrix-assisted laser desorption/ionization mass spectrometry to image host-microbe symbioses and their metabolic interactions. The metaFISH pipeline aligns and integrates metabolite and fluorescent images at the micrometre scale to provide a spatial assignment of host and symbiont metabolites on the same tissue section. To illustrate the advantages of metaFISH, we mapped the spatial metabolome of a deep-sea mussel and its intracellular symbiotic bacteria at the scale of individual epithelial host cells. Our analytical pipeline revealed metabolic adaptations of the epithelial cells to the intracellular symbionts and variation in metabolic phenotypes within a single symbiont 16S rRNA phylotype, and enabled the discovery of specialized metabolites from the host-microbe interface. metaFISH provides a culture-independent approach to link metabolic phenotypes to community members in situ and is a powerful tool for microbiologists across fields.
中文翻译:

微米尺度原位宿主-微生物相互作用的空间代谢组学。
空间代谢组学描述了自然群落中参与代谢表型、防御分子和化学相互作用的小分子的位置和化学性质。目前大多数技术无法在与微生物相互作用相关的尺度上将微生物的基因型和代谢表型原位空间联系起来。在这里,我们提出了一种空间代谢组学流程(metaFISH),它将荧光原位杂交(FISH)显微镜和高分辨率大气压基质辅助激光解吸/电离质谱结合起来,对宿主-微生物共生及其代谢相互作用进行成像。metaFISH 管道在微米尺度上对齐并集成代谢物和荧光图像,以提供同一组织切片上宿主和共生体代谢物的空间分配。为了说明metaFISH的优势,我们在单个上皮宿主细胞的尺度上绘制了深海贻贝及其细胞内共生细菌的空间代谢组图。我们的分析流程揭示了上皮细胞对细胞内共生体的代谢适应以及单一共生体 16S rRNA 系统发育型内代谢表型的变化,并使得能够从宿主-微生物界面发现专门的代谢物。metaFISH 提供了一种独立于培养物的方法,将代谢表型与群落成员原位联系起来,是跨领域微生物学家的强大工具。
更新日期:2020-02-03
中文翻译:

微米尺度原位宿主-微生物相互作用的空间代谢组学。
空间代谢组学描述了自然群落中参与代谢表型、防御分子和化学相互作用的小分子的位置和化学性质。目前大多数技术无法在与微生物相互作用相关的尺度上将微生物的基因型和代谢表型原位空间联系起来。在这里,我们提出了一种空间代谢组学流程(metaFISH),它将荧光原位杂交(FISH)显微镜和高分辨率大气压基质辅助激光解吸/电离质谱结合起来,对宿主-微生物共生及其代谢相互作用进行成像。metaFISH 管道在微米尺度上对齐并集成代谢物和荧光图像,以提供同一组织切片上宿主和共生体代谢物的空间分配。为了说明metaFISH的优势,我们在单个上皮宿主细胞的尺度上绘制了深海贻贝及其细胞内共生细菌的空间代谢组图。我们的分析流程揭示了上皮细胞对细胞内共生体的代谢适应以及单一共生体 16S rRNA 系统发育型内代谢表型的变化,并使得能够从宿主-微生物界面发现专门的代谢物。metaFISH 提供了一种独立于培养物的方法,将代谢表型与群落成员原位联系起来,是跨领域微生物学家的强大工具。



























 京公网安备 11010802027423号
京公网安备 11010802027423号