当前位置:
X-MOL 学术
›
Nat. Rev. Mol. Cell Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Regulatory R-loops as facilitators of gene expression and genome stability.
Nature Reviews Molecular Cell Biology ( IF 81.3 ) Pub Date : 2020-01-31 , DOI: 10.1038/s41580-019-0206-3 Christof Niehrs 1, 2 , Brian Luke 1, 3
Nature Reviews Molecular Cell Biology ( IF 81.3 ) Pub Date : 2020-01-31 , DOI: 10.1038/s41580-019-0206-3 Christof Niehrs 1, 2 , Brian Luke 1, 3
Affiliation
|
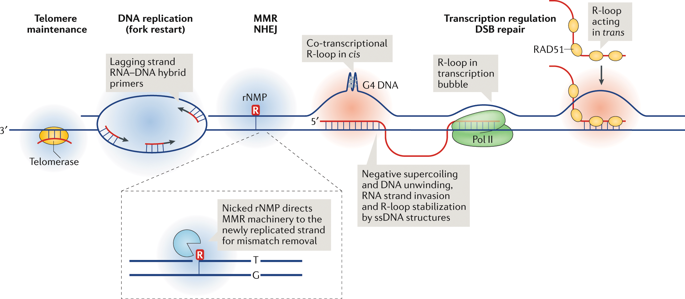
R-loops are three-stranded structures that harbour an RNA-DNA hybrid and frequently form during transcription. R-loop misregulation is associated with DNA damage, transcription elongation defects, hyper-recombination and genome instability. In contrast to such 'unscheduled' R-loops, evidence is mounting that cells harness the presence of RNA-DNA hybrids in scheduled, 'regulatory' R-loops to promote DNA transactions, including transcription termination and other steps of gene regulation, telomere stability and DNA repair. R-loops formed by cellular RNAs can regulate histone post-translational modification and may be recognized by dedicated reader proteins. The two-faced nature of R-loops implies that their formation, location and timely removal must be tightly regulated. In this Perspective, we discuss the cellular processes that regulatory R-loops modulate, the regulation of R-loops and the potential differences that may exist between regulatory R-loops and unscheduled R-loops.
中文翻译:

调节 R 环作为基因表达和基因组稳定性的促进剂。
R 环是三链结构,含有 RNA-DNA 杂合体,在转录过程中经常形成。R-loop 失调与 DNA 损伤、转录延伸缺陷、超重组和基因组不稳定性有关。与这种“计划外”的 R 环相比,越来越多的证据表明,细胞利用预定的“调节性”R 环中存在的 RNA-DNA 杂合体来促进 DNA 交易,包括转录终止和基因调控的其他步骤、端粒稳定性和DNA修复。由细胞 RNA 形成的 R 环可以调节组蛋白的翻译后修饰,并且可以被专门的阅读蛋白识别。R 环的两面性意味着它们的形成、位置和及时去除必须受到严格监管。在这个观点中,
更新日期:2020-01-31
中文翻译:

调节 R 环作为基因表达和基因组稳定性的促进剂。
R 环是三链结构,含有 RNA-DNA 杂合体,在转录过程中经常形成。R-loop 失调与 DNA 损伤、转录延伸缺陷、超重组和基因组不稳定性有关。与这种“计划外”的 R 环相比,越来越多的证据表明,细胞利用预定的“调节性”R 环中存在的 RNA-DNA 杂合体来促进 DNA 交易,包括转录终止和基因调控的其他步骤、端粒稳定性和DNA修复。由细胞 RNA 形成的 R 环可以调节组蛋白的翻译后修饰,并且可以被专门的阅读蛋白识别。R 环的两面性意味着它们的形成、位置和及时去除必须受到严格监管。在这个观点中,











































 京公网安备 11010802027423号
京公网安备 11010802027423号