当前位置:
X-MOL 学术
›
npj Syst. Biol. Appl.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
An integrated computational and experimental study to investigate Staphylococcus aureus metabolism.
npj Systems Biology and Applications ( IF 4 ) Pub Date : 2020-01-30 , DOI: 10.1038/s41540-019-0122-3 Mohammad Mazharul Islam 1 , Vinai C Thomas 2 , Matthew Van Beek 1 , Jong-Sam Ahn 2 , Abdulelah A Alqarzaee 2 , Chunyi Zhou 2 , Paul D Fey 2 , Kenneth W Bayles 2 , Rajib Saha 1
npj Systems Biology and Applications ( IF 4 ) Pub Date : 2020-01-30 , DOI: 10.1038/s41540-019-0122-3 Mohammad Mazharul Islam 1 , Vinai C Thomas 2 , Matthew Van Beek 1 , Jong-Sam Ahn 2 , Abdulelah A Alqarzaee 2 , Chunyi Zhou 2 , Paul D Fey 2 , Kenneth W Bayles 2 , Rajib Saha 1
Affiliation
|
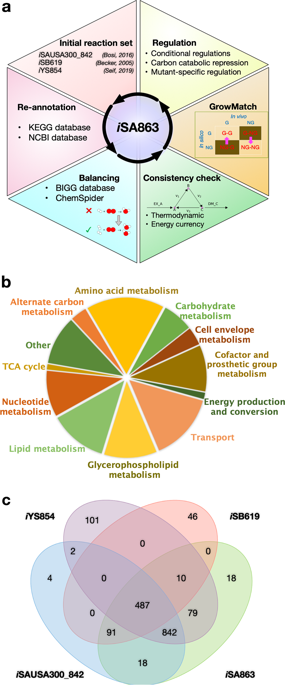
Staphylococcus aureus is a metabolically versatile pathogen that colonizes nearly all organs of the human body. A detailed and comprehensive knowledge of staphylococcal metabolism is essential to understand its pathogenesis. To this end, we have reconstructed and experimentally validated an updated and enhanced genome-scale metabolic model of S. aureus USA300_FPR3757. The model combined genome annotation data, reaction stoichiometry, and regulation information from biochemical databases and previous strain-specific models. Reactions in the model were checked and fixed to ensure chemical balance and thermodynamic consistency. To further refine the model, growth assessment of 1920 nonessential mutants from the Nebraska Transposon Mutant Library was performed, and metabolite excretion profiles of important mutants in carbon and nitrogen metabolism were determined. The growth and no-growth inconsistencies between the model predictions and in vivo essentiality data were resolved using extensive manual curation based on optimization-based reconciliation algorithms. Upon intensive curation and refinements, the model contains 863 metabolic genes, 1379 metabolites (including 1159 unique metabolites), and 1545 reactions including transport and exchange reactions. To improve the accuracy and predictability of the model to environmental changes, condition-specific regulation information curated from the existing knowledgebase was incorporated. These critical additions improved the model performance significantly in capturing gene essentiality, substrate utilization, and metabolite production capabilities and increased the ability to generate model-based discoveries of therapeutic significance. Use of this highly curated model will enhance the functional utility of omics data, and therefore, serve as a resource to support future investigations of S. aureus and to augment staphylococcal research worldwide.
中文翻译:

一项研究金黄色葡萄球菌代谢的综合计算和实验研究。
金黄色葡萄球菌是一种代谢多变的病原体,几乎可以寄居人体的所有器官。详细而全面的葡萄球菌代谢知识对于了解其发病机制至关重要。为此,我们重建并实验验证了金黄色葡萄球菌 USA300_FPR3757 的更新和增强的基因组规模代谢模型。该模型结合了来自生化数据库和先前菌株特异性模型的基因组注释数据、反应化学计量和调节信息。检查并固定模型中的反应以确保化学平衡和热力学一致性。为了进一步完善模型,对来自内布拉斯加州转座子突变体库的 1920 个非必需突变体进行了生长评估,测定了碳和氮代谢中重要突变体的代谢物排泄谱。模型预测和体内重要性数据之间的增长和非增长不一致通过基于优化的协调算法使用广泛的手动管理来解决。经过精心策划和改进,该模型包含 863 个代谢基因、1379 个代谢物(包括 1159 个独特代谢物)和 1545 个反应,包括运输和交换反应。为了提高模型对环境变化的准确性和可预测性,结合了从现有知识库中收集的特定条件的法规信息。这些关键添加显着提高了模型在捕获基因重要性、底物利用率、和代谢物生产能力,并提高了产生具有治疗意义的基于模型的发现的能力。使用这个高度策划的模型将增强组学数据的功能效用,因此,作为支持金黄色葡萄球菌未来研究和加强全球葡萄球菌研究的资源。
更新日期:2020-01-30
中文翻译:

一项研究金黄色葡萄球菌代谢的综合计算和实验研究。
金黄色葡萄球菌是一种代谢多变的病原体,几乎可以寄居人体的所有器官。详细而全面的葡萄球菌代谢知识对于了解其发病机制至关重要。为此,我们重建并实验验证了金黄色葡萄球菌 USA300_FPR3757 的更新和增强的基因组规模代谢模型。该模型结合了来自生化数据库和先前菌株特异性模型的基因组注释数据、反应化学计量和调节信息。检查并固定模型中的反应以确保化学平衡和热力学一致性。为了进一步完善模型,对来自内布拉斯加州转座子突变体库的 1920 个非必需突变体进行了生长评估,测定了碳和氮代谢中重要突变体的代谢物排泄谱。模型预测和体内重要性数据之间的增长和非增长不一致通过基于优化的协调算法使用广泛的手动管理来解决。经过精心策划和改进,该模型包含 863 个代谢基因、1379 个代谢物(包括 1159 个独特代谢物)和 1545 个反应,包括运输和交换反应。为了提高模型对环境变化的准确性和可预测性,结合了从现有知识库中收集的特定条件的法规信息。这些关键添加显着提高了模型在捕获基因重要性、底物利用率、和代谢物生产能力,并提高了产生具有治疗意义的基于模型的发现的能力。使用这个高度策划的模型将增强组学数据的功能效用,因此,作为支持金黄色葡萄球菌未来研究和加强全球葡萄球菌研究的资源。



























 京公网安备 11010802027423号
京公网安备 11010802027423号