Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding.
The Lancet ( IF 98.4 ) Pub Date : 2020-01-30 , DOI: 10.1016/s0140-6736(20)30251-8 Roujian Lu 1 , Xiang Zhao 1 , Juan Li 2 , Peihua Niu 1 , Bo Yang 3 , Honglong Wu 4 , Wenling Wang 1 , Hao Song 5 , Baoying Huang 1 , Na Zhu 1 , Yuhai Bi 6 , Xuejun Ma 1 , Faxian Zhan 3 , Liang Wang 6 , Tao Hu 2 , Hong Zhou 2 , Zhenhong Hu 7 , Weimin Zhou 1 , Li Zhao 1 , Jing Chen 8 , Yao Meng 1 , Ji Wang 1 , Yang Lin 4 , Jianying Yuan 4 , Zhihao Xie 4 , Jinmin Ma 4 , William J Liu 1 , Dayan Wang 1 , Wenbo Xu 1 , Edward C Holmes 9 , George F Gao 10 , Guizhen Wu 1 , Weijun Chen 4 , Weifeng Shi 11 , Wenjie Tan 12
The Lancet ( IF 98.4 ) Pub Date : 2020-01-30 , DOI: 10.1016/s0140-6736(20)30251-8 Roujian Lu 1 , Xiang Zhao 1 , Juan Li 2 , Peihua Niu 1 , Bo Yang 3 , Honglong Wu 4 , Wenling Wang 1 , Hao Song 5 , Baoying Huang 1 , Na Zhu 1 , Yuhai Bi 6 , Xuejun Ma 1 , Faxian Zhan 3 , Liang Wang 6 , Tao Hu 2 , Hong Zhou 2 , Zhenhong Hu 7 , Weimin Zhou 1 , Li Zhao 1 , Jing Chen 8 , Yao Meng 1 , Ji Wang 1 , Yang Lin 4 , Jianying Yuan 4 , Zhihao Xie 4 , Jinmin Ma 4 , William J Liu 1 , Dayan Wang 1 , Wenbo Xu 1 , Edward C Holmes 9 , George F Gao 10 , Guizhen Wu 1 , Weijun Chen 4 , Weifeng Shi 11 , Wenjie Tan 12
Affiliation
|
BACKGROUND
In late December, 2019, patients presenting with viral pneumonia due to an unidentified microbial agent were reported in Wuhan, China. A novel coronavirus was subsequently identified as the causative pathogen, provisionally named 2019 novel coronavirus (2019-nCoV). As of Jan 26, 2020, more than 2000 cases of 2019-nCoV infection have been confirmed, most of which involved people living in or visiting Wuhan, and human-to-human transmission has been confirmed.
METHODS
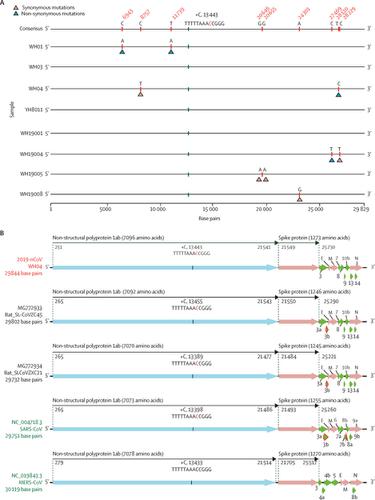
We did next-generation sequencing of samples from bronchoalveolar lavage fluid and cultured isolates from nine inpatients, eight of whom had visited the Huanan seafood market in Wuhan. Complete and partial 2019-nCoV genome sequences were obtained from these individuals. Viral contigs were connected using Sanger sequencing to obtain the full-length genomes, with the terminal regions determined by rapid amplification of cDNA ends. Phylogenetic analysis of these 2019-nCoV genomes and those of other coronaviruses was used to determine the evolutionary history of the virus and help infer its likely origin. Homology modelling was done to explore the likely receptor-binding properties of the virus.
FINDINGS
The ten genome sequences of 2019-nCoV obtained from the nine patients were extremely similar, exhibiting more than 99·98% sequence identity. Notably, 2019-nCoV was closely related (with 88% identity) to two bat-derived severe acute respiratory syndrome (SARS)-like coronaviruses, bat-SL-CoVZC45 and bat-SL-CoVZXC21, collected in 2018 in Zhoushan, eastern China, but were more distant from SARS-CoV (about 79%) and MERS-CoV (about 50%). Phylogenetic analysis revealed that 2019-nCoV fell within the subgenus Sarbecovirus of the genus Betacoronavirus, with a relatively long branch length to its closest relatives bat-SL-CoVZC45 and bat-SL-CoVZXC21, and was genetically distinct from SARS-CoV. Notably, homology modelling revealed that 2019-nCoV had a similar receptor-binding domain structure to that of SARS-CoV, despite amino acid variation at some key residues.
INTERPRETATION
2019-nCoV is sufficiently divergent from SARS-CoV to be considered a new human-infecting betacoronavirus. Although our phylogenetic analysis suggests that bats might be the original host of this virus, an animal sold at the seafood market in Wuhan might represent an intermediate host facilitating the emergence of the virus in humans. Importantly, structural analysis suggests that 2019-nCoV might be able to bind to the angiotensin-converting enzyme 2 receptor in humans. The future evolution, adaptation, and spread of this virus warrant urgent investigation.
FUNDING
National Key Research and Development Program of China, National Major Project for Control and Prevention of Infectious Disease in China, Chinese Academy of Sciences, Shandong First Medical University.
更新日期:2020-02-21











































 京公网安备 11010802027423号
京公网安备 11010802027423号