当前位置:
X-MOL 学术
›
J. Biomed. Inform.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Combinatorial feature embedding based on CNN and LSTM for biomedical named entity recognition.
Journal of Biomedical informatics ( IF 4.0 ) Pub Date : 2020-01-28 , DOI: 10.1016/j.jbi.2020.103381 Minsoo Cho 1 , Jihwan Ha 1 , Chihyun Park 1 , Sanghyun Park 1
Journal of Biomedical informatics ( IF 4.0 ) Pub Date : 2020-01-28 , DOI: 10.1016/j.jbi.2020.103381 Minsoo Cho 1 , Jihwan Ha 1 , Chihyun Park 1 , Sanghyun Park 1
Affiliation
|
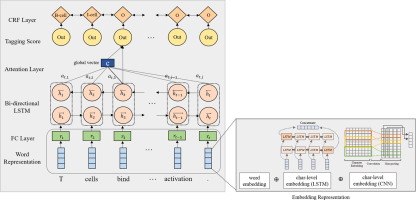
With the rapid advancement of technology and the necessity of processing large amounts of data, biomedical Named Entity Recognition (NER) has become an essential technique for information extraction in the biomedical field. NER, which is a sequence-labeling task, has been performed using various traditional techniques including dictionary-, rule-, machine learning-, and deep learning-based methods. However, as existing biomedical NER models are insufficient to handle new and unseen entity types from the growing biomedical data, the development of more effective and accurate biomedical NER models is being widely researched. Among biomedical NER models utilizing deep learning approaches, there have been only a few studies involving the design of high-level features in the embedding layer. In this regard, herein, we propose a deep learning NER model that effectively represents biomedical word tokens through the design of a combinatorial feature embedding. The proposed model is based on Bidirectional Long Short-Term Memory (bi-LSTM) with Conditional Random Field (CRF) and enhanced by integrating two different character-level representations extracted from a Convolutional Neural Network (CNN) and bi-LSTM. Additionally, an attention mechanism is applied to the model to focus on the relevant tokens in the sentence, which alleviates the long-term dependency problem of the LSTM model and allows effective recognition of entities. The proposed model was evaluated on two benchmark datasets, the JNLPBA and NCBI-Disease, and a comparative analysis with the existing models is performed. The proposed model achieved a relatively higher performance with an F1-score of 86.93% in case of NCBI-Disease, and a competitive performance for the JNLPBA with an F1-score of 75.31%.
中文翻译:

基于CNN和LSTM的组合特征嵌入用于生物医学命名实体识别。
随着技术的飞速发展和处理大量数据的必要性,生物医学命名实体识别(NER)已成为生物医学领域信息提取的必不可少的技术。NER是一种序列标记任务,已使用各种传统技术执行,包括字典,规则,机器学习和基于深度学习的方法。但是,由于现有的生物医学NER模型不足以处理来自不断增长的生物医学数据的新的和看不见的实体类型,因此人们正在广泛研究开发更有效和准确的生物医学NER模型。在利用深度学习方法的生物医学NER模型中,只有很少的研究涉及嵌入层中高级特征的设计。在这方面,我们提出了一种深度学习NER模型,该模型可通过组合特征嵌入的设计有效地表示生物医学单词标记。所提出的模型基于带有条件随机场(CRF)的双向长短期记忆(bi-LSTM),并通过集成从卷积神经网络(CNN)和bi-LSTM中提取的两个不同字符级表示进行了增强。另外,将注意力机制应用于模型以集中于句子中的相关标记,这减轻了LSTM模型的长期依赖性问题并允许有效识别实体。在两个基准数据集JNLPBA和NCBI-Disease上评估了提出的模型,并与现有模型进行了比较分析。
更新日期:2020-01-29
中文翻译:

基于CNN和LSTM的组合特征嵌入用于生物医学命名实体识别。
随着技术的飞速发展和处理大量数据的必要性,生物医学命名实体识别(NER)已成为生物医学领域信息提取的必不可少的技术。NER是一种序列标记任务,已使用各种传统技术执行,包括字典,规则,机器学习和基于深度学习的方法。但是,由于现有的生物医学NER模型不足以处理来自不断增长的生物医学数据的新的和看不见的实体类型,因此人们正在广泛研究开发更有效和准确的生物医学NER模型。在利用深度学习方法的生物医学NER模型中,只有很少的研究涉及嵌入层中高级特征的设计。在这方面,我们提出了一种深度学习NER模型,该模型可通过组合特征嵌入的设计有效地表示生物医学单词标记。所提出的模型基于带有条件随机场(CRF)的双向长短期记忆(bi-LSTM),并通过集成从卷积神经网络(CNN)和bi-LSTM中提取的两个不同字符级表示进行了增强。另外,将注意力机制应用于模型以集中于句子中的相关标记,这减轻了LSTM模型的长期依赖性问题并允许有效识别实体。在两个基准数据集JNLPBA和NCBI-Disease上评估了提出的模型,并与现有模型进行了比较分析。











































 京公网安备 11010802027423号
京公网安备 11010802027423号