Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Machine Learning and Network Analyses Reveal Disease Subtypes of Pancreatic Cancer and their Molecular Characteristics.
Scientific Reports ( IF 3.8 ) Pub Date : 2020-01-27 , DOI: 10.1038/s41598-020-58290-2 Musalula Sinkala 1 , Nicola Mulder 1 , Darren Martin 1
Scientific Reports ( IF 3.8 ) Pub Date : 2020-01-27 , DOI: 10.1038/s41598-020-58290-2 Musalula Sinkala 1 , Nicola Mulder 1 , Darren Martin 1
Affiliation
|
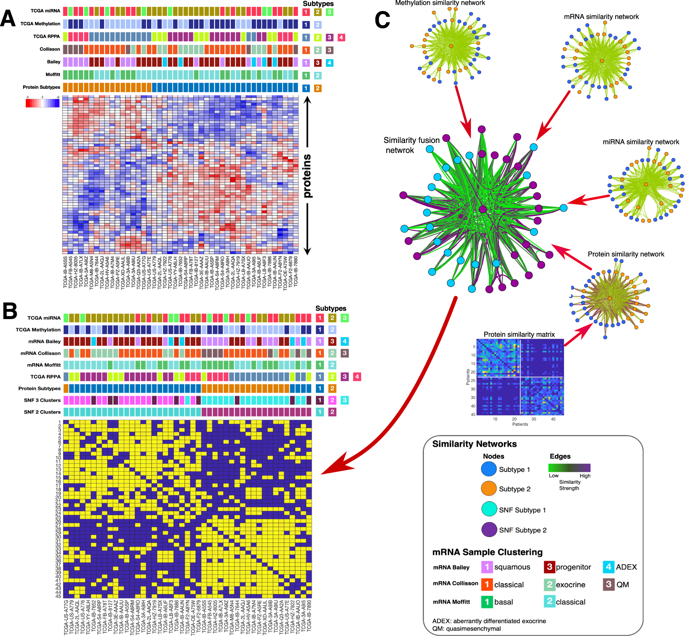
Given that the biological processes governing the oncogenesis of pancreatic cancers could present useful therapeutic targets, there is a pressing need to molecularly distinguish between different clinically relevant pancreatic cancer subtypes. To address this challenge, we used targeted proteomics and other molecular data compiled by The Cancer Genome Atlas to reveal that pancreatic tumours can be broadly segregated into two distinct subtypes. Besides being associated with substantially different clinical outcomes, tumours belonging to each of these subtypes also display notable differences in diverse signalling pathways and biological processes. At the proteome level, we show that tumours belonging to the less severe subtype are characterised by aberrant mTOR signalling, whereas those belonging to the more severe subtype are characterised by disruptions in SMAD and cell cycle-related processes. We use machine learning algorithms to define sets of proteins, mRNAs, miRNAs and DNA methylation patterns that could serve as biomarkers to accurately differentiate between the two pancreatic cancer subtypes. Lastly, we confirm the biological relevance of the identified biomarkers by showing that these can be used together with pattern-recognition algorithms to accurately infer the drug sensitivity of pancreatic cancer cell lines. Our study shows that integrative profiling of multiple data types enables a biological and clinical representation of pancreatic cancer that is comprehensive enough to provide a foundation for future therapeutic strategies.
中文翻译:

机器学习和网络分析揭示胰腺癌的疾病亚型及其分子特征。
鉴于控制胰腺癌肿瘤发生的生物过程可能提供有用的治疗靶点,迫切需要从分子角度区分不同的临床相关胰腺癌亚型。为了应对这一挑战,我们使用癌症基因组图谱编制的靶向蛋白质组学和其他分子数据来揭示胰腺肿瘤可以大致分为两种不同的亚型。除了与显着不同的临床结果相关之外,属于这些亚型的肿瘤在不同的信号通路和生物过程中也表现出显着的差异。在蛋白质组水平上,我们发现属于不太严重的亚型的肿瘤以异常的 mTOR 信号传导为特征,而属于较严重的亚型的肿瘤的特征是 SMAD 和细胞周期相关过程的破坏。我们使用机器学习算法来定义一组蛋白质、mRNA、miRNA 和 DNA 甲基化模式,这些模式可以作为生物标志物来准确区分两种胰腺癌亚型。最后,我们通过证明这些生物标志物可以与模式识别算法一起使用来准确推断胰腺癌细胞系的药物敏感性,从而确认了所识别的生物标志物的生物学相关性。我们的研究表明,多种数据类型的综合分析能够提供足够全面的胰腺癌生物学和临床表征,为未来的治疗策略奠定基础。
更新日期:2020-01-27
中文翻译:

机器学习和网络分析揭示胰腺癌的疾病亚型及其分子特征。
鉴于控制胰腺癌肿瘤发生的生物过程可能提供有用的治疗靶点,迫切需要从分子角度区分不同的临床相关胰腺癌亚型。为了应对这一挑战,我们使用癌症基因组图谱编制的靶向蛋白质组学和其他分子数据来揭示胰腺肿瘤可以大致分为两种不同的亚型。除了与显着不同的临床结果相关之外,属于这些亚型的肿瘤在不同的信号通路和生物过程中也表现出显着的差异。在蛋白质组水平上,我们发现属于不太严重的亚型的肿瘤以异常的 mTOR 信号传导为特征,而属于较严重的亚型的肿瘤的特征是 SMAD 和细胞周期相关过程的破坏。我们使用机器学习算法来定义一组蛋白质、mRNA、miRNA 和 DNA 甲基化模式,这些模式可以作为生物标志物来准确区分两种胰腺癌亚型。最后,我们通过证明这些生物标志物可以与模式识别算法一起使用来准确推断胰腺癌细胞系的药物敏感性,从而确认了所识别的生物标志物的生物学相关性。我们的研究表明,多种数据类型的综合分析能够提供足够全面的胰腺癌生物学和临床表征,为未来的治疗策略奠定基础。











































 京公网安备 11010802027423号
京公网安备 11010802027423号