Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Depicting the genetic architecture of pediatric cancers through an integrative gene network approach.
Scientific Reports ( IF 3.8 ) Pub Date : 2020-01-27 , DOI: 10.1038/s41598-020-58179-0 Clara Savary 1 , Artem Kim 1 , Alexandra Lespagnol 2 , Virginie Gandemer 1, 3 , Isabelle Pellier 4 , Charlotte Andrieu 5, 6 , Gilles Pagès 7, 8 , Marie-Dominique Galibert 1, 2 , Yuna Blum 9 , Marie de Tayrac 1, 5
Scientific Reports ( IF 3.8 ) Pub Date : 2020-01-27 , DOI: 10.1038/s41598-020-58179-0 Clara Savary 1 , Artem Kim 1 , Alexandra Lespagnol 2 , Virginie Gandemer 1, 3 , Isabelle Pellier 4 , Charlotte Andrieu 5, 6 , Gilles Pagès 7, 8 , Marie-Dominique Galibert 1, 2 , Yuna Blum 9 , Marie de Tayrac 1, 5
Affiliation
|
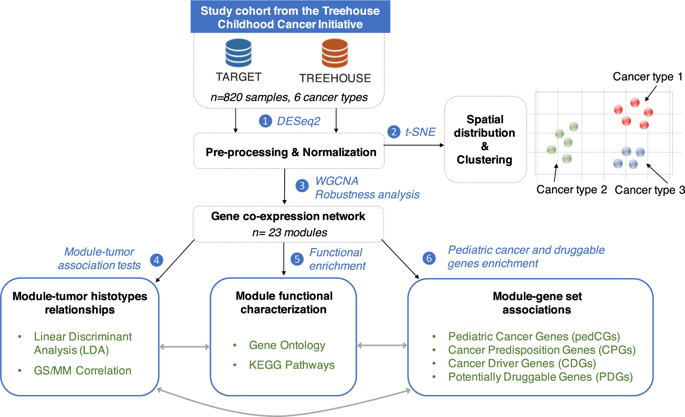
The genetic etiology of childhood cancers still remains largely unknown. It is therefore essential to develop novel strategies to unravel the spectrum of pediatric cancer genes. Statistical network modeling techniques have emerged as powerful methodologies for enabling the inference of gene-disease relationship and have been performed on adult but not pediatric cancers. We performed a deep multi-layer understanding of pan-cancer transcriptome data selected from the Treehouse Childhood Cancer Initiative through a co-expression network analysis. We identified six modules strongly associated with pediatric tumor histotypes that were functionally linked to developmental processes. Topological analyses highlighted that pediatric cancer predisposition genes and potential therapeutic targets were central regulators of cancer-histotype specific modules. A module was related to multiple pediatric malignancies with functions involved in DNA repair and cell cycle regulation. This canonical oncogenic module gathered most of the childhood cancer predisposition genes and clinically actionable genes. In pediatric acute leukemias, the driver genes were co-expressed in a module related to epigenetic and post-transcriptional processes, suggesting a critical role of these pathways in the progression of hematologic malignancies. This integrative pan-cancer study provides a thorough characterization of pediatric tumor-associated modules and paves the way for investigating novel candidate genes involved in childhood tumorigenesis.
中文翻译:

通过综合基因网络方法描述儿童癌症的遗传结构。
儿童癌症的遗传病因学仍然很大程度上未知。因此,开发新颖的策略以揭示儿童癌症基因的谱图至关重要。统计网络建模技术已成为一种强大的方法,可以推断基因-疾病的关系,并且已在成人而非儿科癌症上进行。我们通过共同表达网络分析对从Treehouse Childhood Cancer Initiative中选择的泛癌转录组数据进行了深入的多层了解。我们确定了与儿科肿瘤组织学类型密切相关的六个模块,这些模块在功能上与发育过程相关。拓扑分析强调,儿科癌症的易感基因和潜在的治疗靶点是癌症组织型特定模块的主要调节因子。一个模块与多个儿科恶性肿瘤有关,其功能涉及DNA修复和细胞周期调控。这个典型的致癌模块收集了大多数儿童期癌症易感基因和临床上可操作的基因。在小儿急性白血病中,驱动基因在与表观遗传和转录后过程相关的模块中共表达,提示这些途径在血液系统恶性肿瘤的发展中起着至关重要的作用。这项全面的泛癌研究提供了与儿科肿瘤相关模块的全面表征,为研究与儿童期肿瘤发生有关的新型候选基因铺平了道路。这个典型的致癌模块收集了大多数儿童期癌症易感基因和临床上可操作的基因。在小儿急性白血病中,驱动基因在与表观遗传和转录后过程相关的模块中共表达,提示这些途径在血液系统恶性肿瘤的发展中起着至关重要的作用。这项全面的泛癌研究提供了与儿科肿瘤相关模块的全面表征,为研究与儿童期肿瘤发生有关的新型候选基因铺平了道路。这个典型的致癌模块收集了大多数儿童期癌症易感基因和临床上可操作的基因。在小儿急性白血病中,驱动基因在与表观遗传和转录后过程相关的模块中共表达,提示这些途径在血液系统恶性肿瘤的发展中起着至关重要的作用。这项全面的泛癌研究提供了与儿科肿瘤相关模块的全面表征,为研究与儿童期肿瘤发生有关的新型候选基因铺平了道路。
更新日期:2020-01-27
中文翻译:

通过综合基因网络方法描述儿童癌症的遗传结构。
儿童癌症的遗传病因学仍然很大程度上未知。因此,开发新颖的策略以揭示儿童癌症基因的谱图至关重要。统计网络建模技术已成为一种强大的方法,可以推断基因-疾病的关系,并且已在成人而非儿科癌症上进行。我们通过共同表达网络分析对从Treehouse Childhood Cancer Initiative中选择的泛癌转录组数据进行了深入的多层了解。我们确定了与儿科肿瘤组织学类型密切相关的六个模块,这些模块在功能上与发育过程相关。拓扑分析强调,儿科癌症的易感基因和潜在的治疗靶点是癌症组织型特定模块的主要调节因子。一个模块与多个儿科恶性肿瘤有关,其功能涉及DNA修复和细胞周期调控。这个典型的致癌模块收集了大多数儿童期癌症易感基因和临床上可操作的基因。在小儿急性白血病中,驱动基因在与表观遗传和转录后过程相关的模块中共表达,提示这些途径在血液系统恶性肿瘤的发展中起着至关重要的作用。这项全面的泛癌研究提供了与儿科肿瘤相关模块的全面表征,为研究与儿童期肿瘤发生有关的新型候选基因铺平了道路。这个典型的致癌模块收集了大多数儿童期癌症易感基因和临床上可操作的基因。在小儿急性白血病中,驱动基因在与表观遗传和转录后过程相关的模块中共表达,提示这些途径在血液系统恶性肿瘤的发展中起着至关重要的作用。这项全面的泛癌研究提供了与儿科肿瘤相关模块的全面表征,为研究与儿童期肿瘤发生有关的新型候选基因铺平了道路。这个典型的致癌模块收集了大多数儿童期癌症易感基因和临床上可操作的基因。在小儿急性白血病中,驱动基因在与表观遗传和转录后过程相关的模块中共表达,提示这些途径在血液系统恶性肿瘤的发展中起着至关重要的作用。这项全面的泛癌研究提供了与儿科肿瘤相关模块的全面表征,为研究与儿童期肿瘤发生有关的新型候选基因铺平了道路。











































 京公网安备 11010802027423号
京公网安备 11010802027423号