当前位置:
X-MOL 学术
›
Cell Death Differ.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Deep profiling of apoptotic pathways with mass cytometry identifies a synergistic drug combination for killing myeloma cells.
Cell Death and Differentiation ( IF 13.7 ) Pub Date : 2020-01-27 , DOI: 10.1038/s41418-020-0498-z Charis E Teh 1, 2 , Jia-Nan Gong 1, 2 , David Segal 1, 2 , Tania Tan 1 , Cassandra J Vandenberg 1, 2 , Pasquale L Fedele 1, 2, 3 , Michael S Y Low 1, 2, 3 , George Grigoriadis 3, 4, 5 , Simon J Harrison 6, 7 , Andreas Strasser 1, 2 , Andrew W Roberts 1, 2, 6, 7 , David C S Huang 1, 2 , Garry P Nolan 8, 9 , Daniel H D Gray 1, 2 , Melissa E Ko 8, 9
Cell Death and Differentiation ( IF 13.7 ) Pub Date : 2020-01-27 , DOI: 10.1038/s41418-020-0498-z Charis E Teh 1, 2 , Jia-Nan Gong 1, 2 , David Segal 1, 2 , Tania Tan 1 , Cassandra J Vandenberg 1, 2 , Pasquale L Fedele 1, 2, 3 , Michael S Y Low 1, 2, 3 , George Grigoriadis 3, 4, 5 , Simon J Harrison 6, 7 , Andreas Strasser 1, 2 , Andrew W Roberts 1, 2, 6, 7 , David C S Huang 1, 2 , Garry P Nolan 8, 9 , Daniel H D Gray 1, 2 , Melissa E Ko 8, 9
Affiliation
|
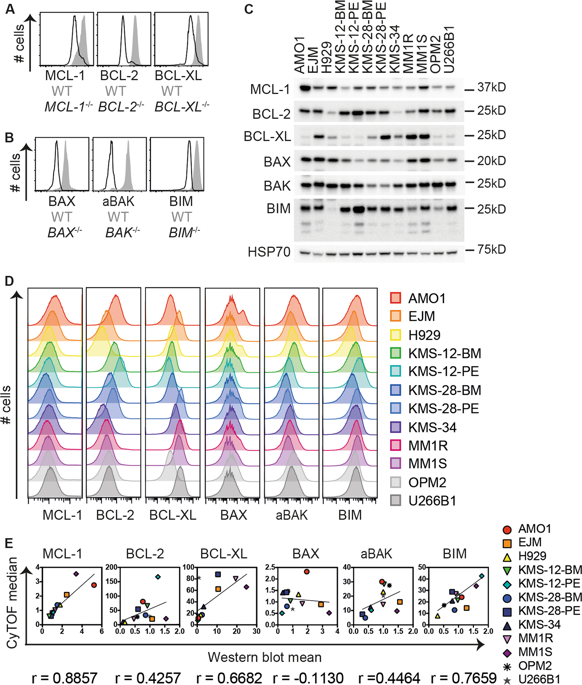
Multiple myeloma is an incurable and fatal cancer of immunoglobulin-secreting plasma cells. Most conventional therapies aim to induce apoptosis in myeloma cells but resistance to these drugs often arises and drives relapse. In this study, we sought to identify the best adjunct targets to kill myeloma cells resistant to conventional therapies using deep profiling by mass cytometry (CyTOF). We validated probes to simultaneously detect 26 regulators of cell death, mitosis, cell signaling, and cancer-related pathways at the single-cell level following treatment of myeloma cells with dexamethasone or bortezomib. Time-resolved visualization algorithms and machine learning random forest models (RFMs) delineated putative cell death trajectories and a hierarchy of parameters that specified myeloma cell survival versus apoptosis following treatment. Among these parameters, increased amounts of phosphorylated cAMP response element-binding protein (CREB) and the pro-survival protein, MCL-1, were defining features of cells surviving drug treatment. Importantly, the RFM prediction that the combination of an MCL-1 inhibitor with dexamethasone would elicit potent, synergistic killing of myeloma cells was validated in other cell lines, in vivo preclinical models and primary myeloma samples from patients. Furthermore, CyTOF analysis of patient bone marrow cells clearly identified myeloma cells and their key cell survival features. This study demonstrates the utility of CyTOF profiling at the single-cell level to identify clinically relevant drug combinations and tracking of patient responses for future clinical trials.
中文翻译:

通过质谱流式细胞仪对细胞凋亡途径进行深入分析,确定了一种用于杀死骨髓瘤细胞的协同药物组合。
多发性骨髓瘤是一种无法治愈且致命的免疫球蛋白分泌浆细胞癌症。大多数传统疗法旨在诱导骨髓瘤细胞凋亡,但通常会出现对这些药物的耐药性并导致复发。在这项研究中,我们试图通过质谱流式分析 (CyTOF) 进行深度分析来确定杀死对传统疗法耐药的骨髓瘤细胞的最佳辅助靶点。我们验证了探针,在用地塞米松或硼替佐米治疗骨髓瘤细胞后,可以在单细胞水平上同时检测细胞死亡、有丝分裂、细胞信号传导和癌症相关途径的 26 个调节因子。时间分辨可视化算法和机器学习随机森林模型 (RFM) 描绘了假定的细胞死亡轨迹和指定骨髓瘤细胞在治疗后存活与凋亡的参数层次结构。在这些参数中,磷酸化 cAMP 反应元件结合蛋白 (CREB) 和促存活蛋白 MCL-1 含量的增加是药物治疗后细胞存活的定义特征。重要的是,RFM 预测 MCL-1 抑制剂与地塞米松的组合将引发对骨髓瘤细胞的有效、协同杀伤,这一预测在其他细胞系、体内临床前模型和患者的原发性骨髓瘤样本中得到了验证。此外,对患者骨髓细胞的 CyTOF 分析清楚地识别了骨髓瘤细胞及其关键的细胞存活特征。这项研究证明了 CyTOF 分析在单细胞水平上的实用性,可用于识别临床相关的药物组合并跟踪患者对未来临床试验的反应。
更新日期:2020-01-27
中文翻译:

通过质谱流式细胞仪对细胞凋亡途径进行深入分析,确定了一种用于杀死骨髓瘤细胞的协同药物组合。
多发性骨髓瘤是一种无法治愈且致命的免疫球蛋白分泌浆细胞癌症。大多数传统疗法旨在诱导骨髓瘤细胞凋亡,但通常会出现对这些药物的耐药性并导致复发。在这项研究中,我们试图通过质谱流式分析 (CyTOF) 进行深度分析来确定杀死对传统疗法耐药的骨髓瘤细胞的最佳辅助靶点。我们验证了探针,在用地塞米松或硼替佐米治疗骨髓瘤细胞后,可以在单细胞水平上同时检测细胞死亡、有丝分裂、细胞信号传导和癌症相关途径的 26 个调节因子。时间分辨可视化算法和机器学习随机森林模型 (RFM) 描绘了假定的细胞死亡轨迹和指定骨髓瘤细胞在治疗后存活与凋亡的参数层次结构。在这些参数中,磷酸化 cAMP 反应元件结合蛋白 (CREB) 和促存活蛋白 MCL-1 含量的增加是药物治疗后细胞存活的定义特征。重要的是,RFM 预测 MCL-1 抑制剂与地塞米松的组合将引发对骨髓瘤细胞的有效、协同杀伤,这一预测在其他细胞系、体内临床前模型和患者的原发性骨髓瘤样本中得到了验证。此外,对患者骨髓细胞的 CyTOF 分析清楚地识别了骨髓瘤细胞及其关键的细胞存活特征。这项研究证明了 CyTOF 分析在单细胞水平上的实用性,可用于识别临床相关的药物组合并跟踪患者对未来临床试验的反应。









































 京公网安备 11010802027423号
京公网安备 11010802027423号