当前位置:
X-MOL 学术
›
Nat. Protoc.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Identifying statistically significant chromatin contacts from Hi-C data with FitHiC2.
Nature Protocols ( IF 13.1 ) Pub Date : 2020-01-24 , DOI: 10.1038/s41596-019-0273-0 Arya Kaul 1, 2 , Sourya Bhattacharyya 3 , Ferhat Ay 3, 4
Nature Protocols ( IF 13.1 ) Pub Date : 2020-01-24 , DOI: 10.1038/s41596-019-0273-0 Arya Kaul 1, 2 , Sourya Bhattacharyya 3 , Ferhat Ay 3, 4
Affiliation
|
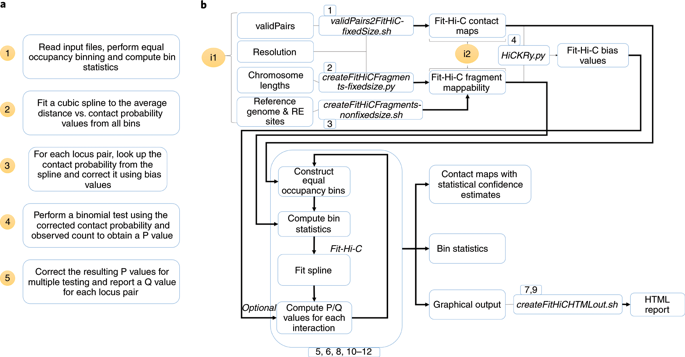
Fit-Hi-C is a programming application to compute statistical confidence estimates for Hi-C contact maps to identify significant chromatin contacts. By fitting a monotonically non-increasing spline, Fit-Hi-C captures the relationship between genomic distance and contact probability without any parametric assumption. The spline fit together with the correction of contact probabilities with respect to bin- or locus-specific biases accounts for previously characterized covariates impacting Hi-C contact counts. Fit-Hi-C is best applied for the study of mid-range (e.g., 20 kb-2 Mb for human genome) intra-chromosomal contacts; however, with the latest reimplementation, named FitHiC2, it is possible to perform genome-wide analysis for high-resolution Hi-C data, including all intra-chromosomal distances and inter-chromosomal contacts. FitHiC2 also offers a merging filter module, which eliminates indirect/bystander interactions, leading to significant reduction in the number of reported contacts without sacrificing recovery of key loops such as those between convergent CTCF binding sites. Here, we describe how to apply the FitHiC2 protocol to three use cases: (i) 5-kb resolution Hi-C data of chromosome 5 from GM12878 (a human lymphoblastoid cell line), (ii) 40-kb resolution whole-genome Hi-C data from IMR90 (human lung fibroblast), and (iii) budding yeast whole-genome Hi-C data at a single restriction cut site (EcoRI) resolution. The procedure takes ~12 h with preprocessing when all use cases are run sequentially (~4 h when run parallel). With the recent improvements in its implementation, FitHiC2 (8 processors and 16 GB memory) is also scalable to genome-wide analysis of the highest resolution (1 kb) Hi-C data available to date (~48 h with 32 GB peak memory). FitHiC2 is available through Bioconda, GitHub and the Python Package Index.
中文翻译:

使用FitHiC2从Hi-C数据中识别出具有统计学意义的染色质接触。
Fit-Hi-C是一种编程应用程序,可为Hi-C联系人图计算统计置信度估计值,以识别重要的染色质联系人。通过拟合单调非递增样条,Fit-Hi-C无需任何参数假设即可捕获基因组距离与接触概率之间的关系。样条拟合以及针对bin或轨迹特定偏差的接触概率校正,共同解释了影响Hi-C接触计数的先前表征的协变量。Fit-Hi-C最适合用于中等范围(例如,人类基因组的20 kb-2 Mb)染色体内接触的研究;但是,使用名为FitHiC2的最新重新实现方法,可以对高分辨率Hi-C数据进行全基因组分析,包括所有染色体内距离和染色体间接触。FitHiC2还提供了合并过滤器模块,该模块消除了间接/旁观者之间的互动,从而在不牺牲关键环(如聚合CTCF绑定位点之间的环)恢复的情况下,大大减少了所报告的联系数量。在这里,我们描述如何将FitHiC2协议应用于三个用例:(i)来自GM12878(人类淋巴母细胞系)的5号染色体的5 kb分辨率Hi-C数据,(ii)40 kb分辨率的全基因组Hi来自IMR90(人肺成纤维细胞)的-C数据,以及(iii)在单个限制性酶切位点(EcoRI)分辨率下出芽的酵母全基因组Hi-C数据。当所有用例依次运行时,该过程需要约12小时的预处理(并行运行时约为4小时)。随着其实施方面的最新改进,FitHiC2(8个处理器和16 GB内存)还可以扩展到全基因组分析,以分析迄今为止可用的最高分辨率(1 kb)Hi-C数据(约48小时,峰值内存为32 GB)。可通过Bioconda,GitHub和Python Package Index获得FitHiC2。
更新日期:2020-01-24
中文翻译:

使用FitHiC2从Hi-C数据中识别出具有统计学意义的染色质接触。
Fit-Hi-C是一种编程应用程序,可为Hi-C联系人图计算统计置信度估计值,以识别重要的染色质联系人。通过拟合单调非递增样条,Fit-Hi-C无需任何参数假设即可捕获基因组距离与接触概率之间的关系。样条拟合以及针对bin或轨迹特定偏差的接触概率校正,共同解释了影响Hi-C接触计数的先前表征的协变量。Fit-Hi-C最适合用于中等范围(例如,人类基因组的20 kb-2 Mb)染色体内接触的研究;但是,使用名为FitHiC2的最新重新实现方法,可以对高分辨率Hi-C数据进行全基因组分析,包括所有染色体内距离和染色体间接触。FitHiC2还提供了合并过滤器模块,该模块消除了间接/旁观者之间的互动,从而在不牺牲关键环(如聚合CTCF绑定位点之间的环)恢复的情况下,大大减少了所报告的联系数量。在这里,我们描述如何将FitHiC2协议应用于三个用例:(i)来自GM12878(人类淋巴母细胞系)的5号染色体的5 kb分辨率Hi-C数据,(ii)40 kb分辨率的全基因组Hi来自IMR90(人肺成纤维细胞)的-C数据,以及(iii)在单个限制性酶切位点(EcoRI)分辨率下出芽的酵母全基因组Hi-C数据。当所有用例依次运行时,该过程需要约12小时的预处理(并行运行时约为4小时)。随着其实施方面的最新改进,FitHiC2(8个处理器和16 GB内存)还可以扩展到全基因组分析,以分析迄今为止可用的最高分辨率(1 kb)Hi-C数据(约48小时,峰值内存为32 GB)。可通过Bioconda,GitHub和Python Package Index获得FitHiC2。











































 京公网安备 11010802027423号
京公网安备 11010802027423号