当前位置:
X-MOL 学术
›
J. Proteome Res.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Comparing 22 Popular Phosphoproteomics Pipelines for Peptide Identification and Site Localization.
Journal of Proteome Research ( IF 3.8 ) Pub Date : 2020-01-24 , DOI: 10.1021/acs.jproteome.9b00679 Marie Locard-Paulet 1, 2 , David Bouyssié 2 , Carine Froment 2 , Odile Burlet-Schiltz 2 , Lars J Jensen 1
Journal of Proteome Research ( IF 3.8 ) Pub Date : 2020-01-24 , DOI: 10.1021/acs.jproteome.9b00679 Marie Locard-Paulet 1, 2 , David Bouyssié 2 , Carine Froment 2 , Odile Burlet-Schiltz 2 , Lars J Jensen 1
Affiliation
|
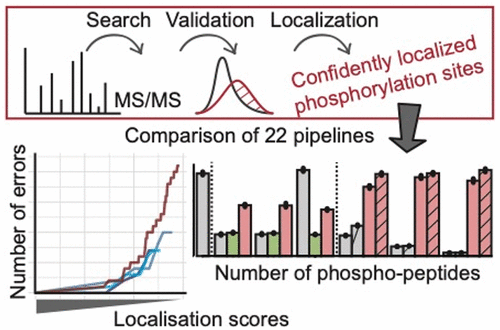
Phosphorylation-driven cell signaling governs most biological functions and is widely studied using mass-spectrometry-based phosphoproteomics. Identifying the peptides and localizing the phosphorylation sites within them from the raw data is challenging and can be performed by several algorithms that return scores that are not directly comparable. This increases the heterogeneity among published phosphoproteomics data sets and prevents their direct integration. Here we compare 22 pipelines implemented in the main software tools used for bottom-up phosphoproteomics analysis (MaxQuant, Proteome Discoverer, PeptideShaker). We test six search engines (Andromeda, Comet, Mascot, MS Amanda, SequestHT, and X!Tandem) in combination with several localization scoring algorithms (delta score, D-score, PTM-score, phosphoRS, and Ascore). We show that these follow very different score distributions, which can lead to different false localization rates for the same threshold. We provide a strategy to discriminate correctly from incorrectly localized phosphorylation sites in a consistent manner across the tested pipelines. The results presented here can help users choose the most appropriate pipeline and cutoffs for their phosphoproteomics analysis.
中文翻译:

比较22条流行的磷酸蛋白组学管线用于肽鉴定和位点定位。
磷酸化驱动的细胞信号传导控制着大多数生物学功能,并且使用基于质谱的磷酸化蛋白质组学进行了广泛的研究。从原始数据中鉴定肽并将其磷酸化位点定位是一项艰巨的任务,可以通过返回不直接可比分的几种算法来执行。这增加了已发表的磷酸蛋白质组学数据集之间的异质性,并阻止了它们的直接整合。在这里,我们比较了用于自下而上的磷酸蛋白质组学分析的主要软件工具(MaxQuant,Proteome Discoverer,PeptideShaker)中实现的22条管道。我们结合几种本地化评分算法(delta评分,D评分,PTM评分,phosphorRS和Ascore)测试了六个搜索引擎(仙女座,彗星,吉祥物,MS Amanda,SequestHT和X!Tandem)。我们表明,这些遵循非常不同的分数分布,对于相同的阈值,这可能导致不同的错误定位率。我们提供了一种策略,可以在测试的管道中以一致的方式正确地区分错误地定位的磷酸化位点。此处显示的结果可帮助用户选择最合适的管线和截止点进行磷酸化蛋白质组学分析。
更新日期:2020-02-06
中文翻译:

比较22条流行的磷酸蛋白组学管线用于肽鉴定和位点定位。
磷酸化驱动的细胞信号传导控制着大多数生物学功能,并且使用基于质谱的磷酸化蛋白质组学进行了广泛的研究。从原始数据中鉴定肽并将其磷酸化位点定位是一项艰巨的任务,可以通过返回不直接可比分的几种算法来执行。这增加了已发表的磷酸蛋白质组学数据集之间的异质性,并阻止了它们的直接整合。在这里,我们比较了用于自下而上的磷酸蛋白质组学分析的主要软件工具(MaxQuant,Proteome Discoverer,PeptideShaker)中实现的22条管道。我们结合几种本地化评分算法(delta评分,D评分,PTM评分,phosphorRS和Ascore)测试了六个搜索引擎(仙女座,彗星,吉祥物,MS Amanda,SequestHT和X!Tandem)。我们表明,这些遵循非常不同的分数分布,对于相同的阈值,这可能导致不同的错误定位率。我们提供了一种策略,可以在测试的管道中以一致的方式正确地区分错误地定位的磷酸化位点。此处显示的结果可帮助用户选择最合适的管线和截止点进行磷酸化蛋白质组学分析。











































 京公网安备 11010802027423号
京公网安备 11010802027423号