当前位置:
X-MOL 学术
›
Nat. Commun.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Transcriptomic profiling of skeletal muscle adaptations to exercise and inactivity.
Nature Communications ( IF 14.7 ) Pub Date : 2020-01-24 , DOI: 10.1038/s41467-019-13869-w Nicolas J Pillon 1 , Brendan M Gabriel 1 , Lucile Dollet 1 , Jonathon A B Smith 1 , Laura Sardón Puig 2 , Javier Botella 3 , David J Bishop 3 , Anna Krook 1 , Juleen R Zierath 1, 2, 4
Nature Communications ( IF 14.7 ) Pub Date : 2020-01-24 , DOI: 10.1038/s41467-019-13869-w Nicolas J Pillon 1 , Brendan M Gabriel 1 , Lucile Dollet 1 , Jonathon A B Smith 1 , Laura Sardón Puig 2 , Javier Botella 3 , David J Bishop 3 , Anna Krook 1 , Juleen R Zierath 1, 2, 4
Affiliation
|
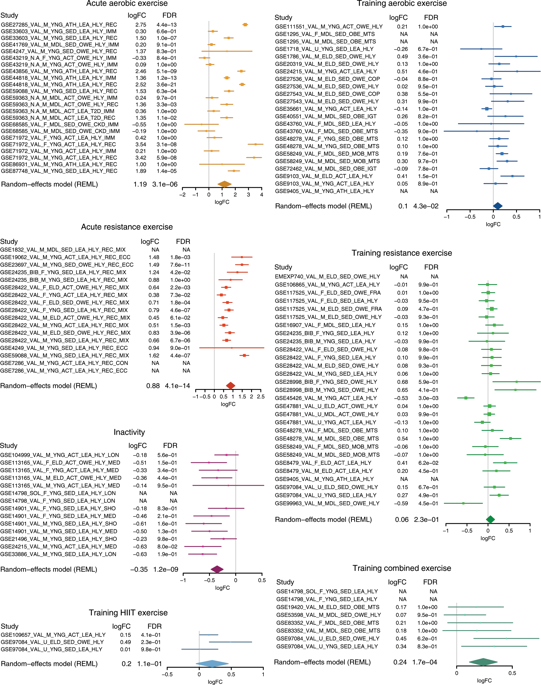
The molecular mechanisms underlying the response to exercise and inactivity are not fully understood. We propose an innovative approach to profile the skeletal muscle transcriptome to exercise and inactivity using 66 published datasets. Data collected from human studies of aerobic and resistance exercise, including acute and chronic exercise training, were integrated using meta-analysis methods (www.metamex.eu). Here we use gene ontology and pathway analyses to reveal selective pathways activated by inactivity, aerobic versus resistance and acute versus chronic exercise training. We identify NR4A3 as one of the most exercise- and inactivity-responsive genes, and establish a role for this nuclear receptor in mediating the metabolic responses to exercise-like stimuli in vitro. The meta-analysis (MetaMEx) also highlights the differential response to exercise in individuals with metabolic impairments. MetaMEx provides the most extensive dataset of skeletal muscle transcriptional responses to different modes of exercise and an online interface to readily interrogate the database.
中文翻译:

骨骼肌适应运动和不活动的转录组分析。
对运动和不运动反应的潜在分子机制尚未完全了解。我们提出了一种创新的方法,使用66个公开的数据集来分析骨骼肌转录组的运动和非活动状态。使用荟萃分析方法(www.metamex.eu)整合了人类对有氧运动和阻力运动研究的数据,包括急性和慢性运动训练。在这里,我们使用基因本体论和途径分析来揭示无活动,有氧与抵抗,急性与慢性运动训练激活的选择性途径。我们将NR4A3确定为对运动和非活动性反应最强的基因之一,并在体外介导对运动样刺激的代谢反应中建立这种核受体的作用。荟萃分析(MetaMEx)还强调了代谢障碍患者对运动的不同反应。MetaMEx提供了最广泛的骨骼肌转录反应对不同运动模式的数据集,并提供了一个易于查询数据库的在线界面。
更新日期:2020-01-24
中文翻译:

骨骼肌适应运动和不活动的转录组分析。
对运动和不运动反应的潜在分子机制尚未完全了解。我们提出了一种创新的方法,使用66个公开的数据集来分析骨骼肌转录组的运动和非活动状态。使用荟萃分析方法(www.metamex.eu)整合了人类对有氧运动和阻力运动研究的数据,包括急性和慢性运动训练。在这里,我们使用基因本体论和途径分析来揭示无活动,有氧与抵抗,急性与慢性运动训练激活的选择性途径。我们将NR4A3确定为对运动和非活动性反应最强的基因之一,并在体外介导对运动样刺激的代谢反应中建立这种核受体的作用。荟萃分析(MetaMEx)还强调了代谢障碍患者对运动的不同反应。MetaMEx提供了最广泛的骨骼肌转录反应对不同运动模式的数据集,并提供了一个易于查询数据库的在线界面。











































 京公网安备 11010802027423号
京公网安备 11010802027423号