当前位置:
X-MOL 学术
›
Nat. Commun.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Integrating multiple genomic technologies to investigate an outbreak of carbapenemase-producing Enterobacter hormaechei.
Nature Communications ( IF 14.7 ) Pub Date : 2020-01-24 , DOI: 10.1038/s41467-019-14139-5 Leah W Roberts 1, 2, 3 , Patrick N A Harris 4, 5 , Brian M Forde 1, 2, 3 , Nouri L Ben Zakour 1, 2, 3 , Elizabeth Catchpoole 5 , Mitchell Stanton-Cook 1, 2 , Minh-Duy Phan 1, 2 , Hanna E Sidjabat 2, 4 , Haakon Bergh 5 , Claire Heney 5 , Jayde A Gawthorne 1, 2 , Jeffrey Lipman 4, 6 , Anthony Allworth 7 , Kok-Gan Chan 8, 9 , Teik Min Chong 8 , Wai-Fong Yin 8 , Mark A Schembri 1, 2 , David L Paterson 2, 4 , Scott A Beatson 1, 2, 3
Nature Communications ( IF 14.7 ) Pub Date : 2020-01-24 , DOI: 10.1038/s41467-019-14139-5 Leah W Roberts 1, 2, 3 , Patrick N A Harris 4, 5 , Brian M Forde 1, 2, 3 , Nouri L Ben Zakour 1, 2, 3 , Elizabeth Catchpoole 5 , Mitchell Stanton-Cook 1, 2 , Minh-Duy Phan 1, 2 , Hanna E Sidjabat 2, 4 , Haakon Bergh 5 , Claire Heney 5 , Jayde A Gawthorne 1, 2 , Jeffrey Lipman 4, 6 , Anthony Allworth 7 , Kok-Gan Chan 8, 9 , Teik Min Chong 8 , Wai-Fong Yin 8 , Mark A Schembri 1, 2 , David L Paterson 2, 4 , Scott A Beatson 1, 2, 3
Affiliation
|
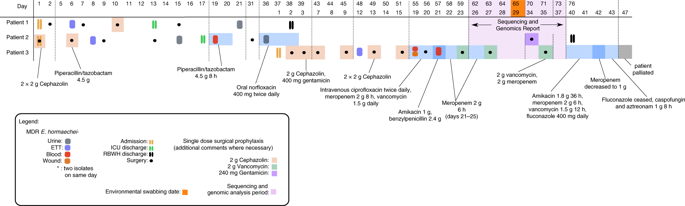
Carbapenem-resistant Enterobacteriaceae (CRE) represent an urgent threat to human health. Here we report the application of several complementary whole-genome sequencing (WGS) technologies to characterise a hospital outbreak of blaIMP-4 carbapenemase-producing E. hormaechei. Using Illumina sequencing, we determined that all outbreak strains were sequence type 90 (ST90) and near-identical. Comparison to publicly available data linked all outbreak isolates to a 2013 isolate from the same ward, suggesting an environmental source in the hospital. Using Pacific Biosciences sequencing, we resolved the complete context of the blaIMP-4 gene on a large IncHI2 plasmid carried by all IMP-4-producing strains across different hospitals. Shotgun metagenomic sequencing of environmental samples also found evidence of ST90 E. hormaechei and the IncHI2 plasmid within the hospital plumbing. Finally, Oxford Nanopore sequencing rapidly resolved the true relationship of subsequent isolates to the initial outbreak. Overall, our strategic application of three WGS technologies provided an in-depth analysis of the outbreak.
中文翻译:

整合了多种基因组技术,调查了产生碳青霉烯酶的霍曼肠杆菌的爆发。
耐碳青霉烯的肠杆菌科(CRE)对人类健康构成紧急威胁。在这里,我们报告了几种互补的全基因组测序(WGS)技术的应用,以表征产生blaIMP-4碳青霉烯酶的霍乱大肠埃希菌的医院暴发。使用Illumina测序,我们确定所有暴发菌株均为序列类型90(ST90),并且几乎相同。与可公开获得的数据进行比较后,将所有疫情隔离株与同一病房的2013年隔离株联系起来,表明医院存在环境污染源。使用Pacific Biosciences测序,我们解析了跨不同医院的所有IMP-4生产菌株携带的大型IncHI2质粒上blaIMP-4基因的完整情况。gun弹枪宏基因组测序的环境样品也发现了ST90 E的证据。医院管道中的Hormaechei和IncHI2质粒。最终,牛津纳米孔测序快速解决了后续分离株与最初暴发之间的真正关系。总体而言,我们对三种WGS技术的战略应用对疫情进行了深入分析。
更新日期:2020-01-24
中文翻译:

整合了多种基因组技术,调查了产生碳青霉烯酶的霍曼肠杆菌的爆发。
耐碳青霉烯的肠杆菌科(CRE)对人类健康构成紧急威胁。在这里,我们报告了几种互补的全基因组测序(WGS)技术的应用,以表征产生blaIMP-4碳青霉烯酶的霍乱大肠埃希菌的医院暴发。使用Illumina测序,我们确定所有暴发菌株均为序列类型90(ST90),并且几乎相同。与可公开获得的数据进行比较后,将所有疫情隔离株与同一病房的2013年隔离株联系起来,表明医院存在环境污染源。使用Pacific Biosciences测序,我们解析了跨不同医院的所有IMP-4生产菌株携带的大型IncHI2质粒上blaIMP-4基因的完整情况。gun弹枪宏基因组测序的环境样品也发现了ST90 E的证据。医院管道中的Hormaechei和IncHI2质粒。最终,牛津纳米孔测序快速解决了后续分离株与最初暴发之间的真正关系。总体而言,我们对三种WGS技术的战略应用对疫情进行了深入分析。











































 京公网安备 11010802027423号
京公网安备 11010802027423号