当前位置:
X-MOL 学术
›
J. Comput. Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Three‐site and five‐site fixed‐charge water models compatible with AMOEBA force field
Journal of Computational Chemistry ( IF 3.4 ) Pub Date : 2020-01-24 , DOI: 10.1002/jcc.26151 Cong Pan 1 , Chengwen Liu 2 , Junhui Peng 1 , Pengyu Ren 2 , Xuhui Huang 1, 3
Journal of Computational Chemistry ( IF 3.4 ) Pub Date : 2020-01-24 , DOI: 10.1002/jcc.26151 Cong Pan 1 , Chengwen Liu 2 , Junhui Peng 1 , Pengyu Ren 2 , Xuhui Huang 1, 3
Affiliation
|
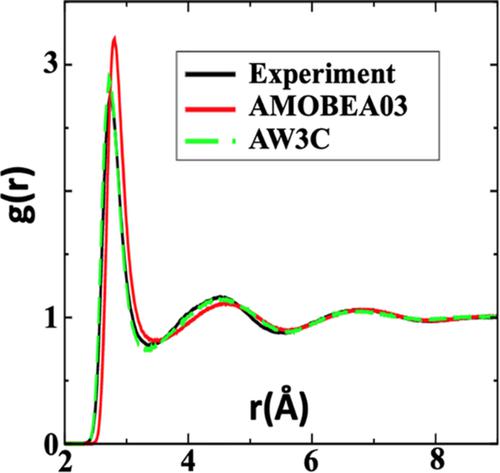
In a typical biomolecular simulation using Atomic Multipole Optimized Energetics for Biomolecular Applications (AMOEBA) force field, the vast majority molecules in the simulation box consist of water, and these water molecules consume the most CPU power due to the explicit mutual induction effect. To improve the computational efficiency, we here develop two new nonpolarizable water models (with flexible bonds and fixed charges) that are compatible with AMOEBA solute: the 3‐site AW3C and 5‐site AW5C. To derive the force‐field parameters for AW3C and AW5C, we fit to six experimental liquid thermodynamic properties: liquid density, enthalpy of vaporization, dielectric constant, isobaric heat capacity, isothermal compressibility and thermal expansion coefficient, at a broad range of temperatures from 261.15 to 353.15 K under 1.0 atm pressure. We further validate our AW3C and AW5C water models by showing that they can well reproduce the radial distribution function g(r), self‐diffusion constant D, and hydration free energy from the AMOEBA03 water model and the experimental observations. Furthermore, we show that our AW3C and AW5C water models can greatly accelerate (>5 times) the bulk water as well as biomolecular simulations when compared to AMOEBA water. Specifically, we demonstrate that the applications of AW3C and AW5C water models to simulate a DNA duplex lead to a threefold acceleration, and in the meanwhile well maintain the structural properties as the fully polarizable AMOEBA water. We expect that our AW3C and AW5C water models hold great promise to be widely applied to simulate complex bio‐molecules using the AMOEBA force field.
中文翻译:

与 AMOEBA 力场兼容的三点和五点固定电荷水模型
在使用 Atomic Multipole Optimized Energetics for Biomolecular Applications (AMOEBA) 力场的典型生物分子模拟中,模拟盒中的绝大多数分子由水组成,由于显式互感应效应,这些水分子消耗了最多的 CPU 功率。为了提高计算效率,我们在这里开发了两种与 AMOEBA 溶质兼容的新的非极化水模型(具有柔性键和固定电荷):3 位点 AW3C 和 5 位点 AW5C。为了推导出 AW3C 和 AW5C 的力场参数,我们拟合了六种实验液体热力学特性:液体密度、汽化焓、介电常数、等压热容、等温压缩率和热膨胀系数,在 261.15 的广泛温度范围内在 1.0 atm 压力下达到 353.15 K。我们进一步验证了我们的 AW3C 和 AW5C 水模型,表明它们可以很好地再现来自 AMOEBA03 水模型和实验观察的径向分布函数 g(r)、自扩散常数 D 和水合自由能。此外,我们表明,与 AMOEBA 水相比,我们的 AW3C 和 AW5C 水模型可以大大加速(> 5 倍)大量水以及生物分子模拟。具体来说,我们证明了应用 AW3C 和 AW5C 水模型来模拟 DNA 双链体导致三倍加速,同时很好地保持了完全可极化的阿米巴水的结构特性。我们希望我们的 AW3C 和 AW5C 水模型有望广泛应用于使用 AMOEBA 力场模拟复杂生物分子。
更新日期:2020-01-24
中文翻译:

与 AMOEBA 力场兼容的三点和五点固定电荷水模型
在使用 Atomic Multipole Optimized Energetics for Biomolecular Applications (AMOEBA) 力场的典型生物分子模拟中,模拟盒中的绝大多数分子由水组成,由于显式互感应效应,这些水分子消耗了最多的 CPU 功率。为了提高计算效率,我们在这里开发了两种与 AMOEBA 溶质兼容的新的非极化水模型(具有柔性键和固定电荷):3 位点 AW3C 和 5 位点 AW5C。为了推导出 AW3C 和 AW5C 的力场参数,我们拟合了六种实验液体热力学特性:液体密度、汽化焓、介电常数、等压热容、等温压缩率和热膨胀系数,在 261.15 的广泛温度范围内在 1.0 atm 压力下达到 353.15 K。我们进一步验证了我们的 AW3C 和 AW5C 水模型,表明它们可以很好地再现来自 AMOEBA03 水模型和实验观察的径向分布函数 g(r)、自扩散常数 D 和水合自由能。此外,我们表明,与 AMOEBA 水相比,我们的 AW3C 和 AW5C 水模型可以大大加速(> 5 倍)大量水以及生物分子模拟。具体来说,我们证明了应用 AW3C 和 AW5C 水模型来模拟 DNA 双链体导致三倍加速,同时很好地保持了完全可极化的阿米巴水的结构特性。我们希望我们的 AW3C 和 AW5C 水模型有望广泛应用于使用 AMOEBA 力场模拟复杂生物分子。











































 京公网安备 11010802027423号
京公网安备 11010802027423号