Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Energy landscape of domain motion in glutamate dehydrogenase deduced from cryo-electron microscopy.
The FEBS Journal ( IF 5.5 ) Pub Date : 2020-01-24 , DOI: 10.1111/febs.15224 Mao Oide 1, 2 , Takayuki Kato 3 , Tomotaka Oroguchi 1, 2 , Masayoshi Nakasako 1, 2
The FEBS Journal ( IF 5.5 ) Pub Date : 2020-01-24 , DOI: 10.1111/febs.15224 Mao Oide 1, 2 , Takayuki Kato 3 , Tomotaka Oroguchi 1, 2 , Masayoshi Nakasako 1, 2
Affiliation
|
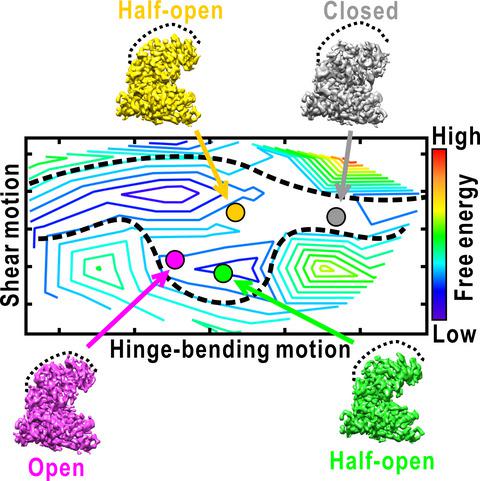
Analysis of the conformational changes of protein is important to elucidate the mechanisms of protein motions correlating with their function. Here, we studied the spontaneous domain motion of unliganded glutamate dehydrogenase from Thermococcus profundus using cryo‐electron microscopy and proposed a novel method to construct free‐energy landscape of protein conformations. Each subunit of the homo‐hexameric enzyme comprises nucleotide‐binding domain (NAD domain) and hexamer‐forming core domain. A large active‐site cleft is situated between the two domains and varies from open to close according to the motion of a NAD domain. A three‐dimensional map reconstructed from all cryo‐electron microscopy images displayed disordered volumes of NAD domains, suggesting that NAD domains in the collected images adopted various conformations in domain motion. Focused classifications on NAD domain of subunits provided several maps of possible conformations in domain motion. To deduce what kinds of conformations appeared in EM images, we developed a novel analysis method that describe the EM maps as a linear combination of representative conformations appearing in a 200‐ns molecular dynamics simulation as reference. The analysis enabled us to estimate the appearance frequencies of the representative conformations, which illustrated a free‐energy landscape in domain motion. In the open/close domain motion, two free‐energy basins hindered the direct transformation from open to closed state. Structure models constructed for representative EM maps in classifications demonstrated the correlation between the energy landscape and conformations in domain motion. Based on the results, the domain motion in glutamate dehydrogenase and the analysis method to visualize conformational changes and free‐energy landscape were discussed.
中文翻译:

从低温电子显微镜推导的谷氨酸脱氢酶中域运动的能量景观。
分析蛋白质的构象变化对于阐明蛋白质运动与其功能相关的机制很重要。在这里,我们研究了深部嗜热球菌的非配体谷氨酸脱氢酶的自发域运动使用低温电子显微镜,并提出了一种新的方法来构建蛋白质构象的自由能态。同源六聚体酶的每个亚基均包含核苷酸结合域(NAD域)和形成六聚体的核心域。一个活跃的裂隙位于两个域之间,并根据NAD域的运动而从打开到关闭变化。从所有冷冻电子显微镜图像重建的三维图显示了无序的NAD域体积,这表明所收集图像中的NAD域在域运动中采用了各种构象。集中在亚基的NAD域上的分类提供了域运动中可能构象的若干图。为了推断EM图像中出现了哪种构象,我们开发了一种新颖的分析方法,将EM图描述为代表200 ns分子动力学模拟中出现的代表性构象的线性组合。分析使我们能够估计代表性构象的出现频率,这说明了域运动中的自由能态。在开/关域运动中,两个自由能盆地阻碍了从开状态到闭状态的直接转换。为分类中的代表性EM映射构建的结构模型证明了能量景观与域运动构象之间的相关性。基于结果,讨论了谷氨酸脱氢酶中的域运动以及可视化构象变化和自由能态的分析方法。
更新日期:2020-01-24
中文翻译:

从低温电子显微镜推导的谷氨酸脱氢酶中域运动的能量景观。
分析蛋白质的构象变化对于阐明蛋白质运动与其功能相关的机制很重要。在这里,我们研究了深部嗜热球菌的非配体谷氨酸脱氢酶的自发域运动使用低温电子显微镜,并提出了一种新的方法来构建蛋白质构象的自由能态。同源六聚体酶的每个亚基均包含核苷酸结合域(NAD域)和形成六聚体的核心域。一个活跃的裂隙位于两个域之间,并根据NAD域的运动而从打开到关闭变化。从所有冷冻电子显微镜图像重建的三维图显示了无序的NAD域体积,这表明所收集图像中的NAD域在域运动中采用了各种构象。集中在亚基的NAD域上的分类提供了域运动中可能构象的若干图。为了推断EM图像中出现了哪种构象,我们开发了一种新颖的分析方法,将EM图描述为代表200 ns分子动力学模拟中出现的代表性构象的线性组合。分析使我们能够估计代表性构象的出现频率,这说明了域运动中的自由能态。在开/关域运动中,两个自由能盆地阻碍了从开状态到闭状态的直接转换。为分类中的代表性EM映射构建的结构模型证明了能量景观与域运动构象之间的相关性。基于结果,讨论了谷氨酸脱氢酶中的域运动以及可视化构象变化和自由能态的分析方法。











































 京公网安备 11010802027423号
京公网安备 11010802027423号