当前位置:
X-MOL 学术
›
Anal. Chim. Acta
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
In silico maturation of affinity and selectivity of DNA aptamers against aflatoxin B1 for biosensor development
Analytica Chimica Acta ( IF 5.7 ) Pub Date : 2020-04-01 , DOI: 10.1016/j.aca.2020.01.045 Maryam Mousivand , Laura Anfossi , Kowsar Bagherzadeh , Nadia Barbero , Amir Mirzadi-Gohari , Mohammad Javan-Nikkhah
Analytica Chimica Acta ( IF 5.7 ) Pub Date : 2020-04-01 , DOI: 10.1016/j.aca.2020.01.045 Maryam Mousivand , Laura Anfossi , Kowsar Bagherzadeh , Nadia Barbero , Amir Mirzadi-Gohari , Mohammad Javan-Nikkhah
|
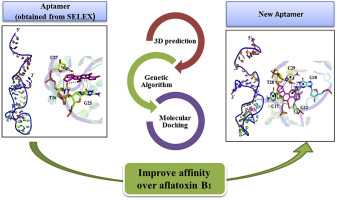
A high affinity and selectivity DNA aptamer for aflatoxin B1 (AFB1) was designed through Genetic Algorithm (GA) based in silico maturation (ISM) strategy. The sequence of a known AFB1 aptamer (Patent: PCT/CA2010/001292, Apt1) applied as a probe in many aptasensors was modified using seven GA rounds to generate an initial library and three different generations of ss DNA oligonucleotides as new candidate aptamers. Molecular docking methodology was used to screen and analyze the best aptamer-AFB1 complexes. Also, a new pipeline was proposed to faithfully predict the tertiary structure of all single stranded DNA sequences. By the second generation, aptamer Apt1 sequence was optimized in the local search space and five aptamers including F20, g12, C52, C32 and H1 were identified as the best aptamers for AFB1. The selected aptamers were applied as probes in an unmodified gold nanoparticles-based aptasensor to evaluate their binding affinity to AFB1 and their selectivity against other mycotoxins (aflatoxins B2, G1, G2, M1, ochratoxin A and zearalenone). In addition, a novel direct fluorescent anisotropy aptamer assay was developed to confirm the binding interaction of the selected aptamers over AFB1. The ISM allowed the identification of an aptamer, F20, with up to 9.4 and 2 fold improvement in affinity and selectivity compared to the parent aptamer, respectively.
中文翻译:

用于生物传感器开发的 DNA 适体对黄曲霉毒素 B1 的亲和力和选择性的计算机成熟
黄曲霉毒素 B1 (AFB1) 的高亲和力和选择性 DNA 适体是通过基于计算机成熟 (ISM) 策略的遗传算法 (GA) 设计的。在许多适体传感器中用作探针的已知 AFB1 适体(专利:PCT/CA2010/001292,Apt1)的序列使用七轮 GA 进行修改,以生成初始文库和三代不同的 ss DNA 寡核苷酸作为新的候选适体。分子对接方法用于筛选和分析最佳适体-AFB1 复合物。此外,还提出了一种新的管道来忠实地预测所有单链 DNA 序列的三级结构。到第二代,适体Apt1序列在局部搜索空间得到优化,F20、g12、C52、C32和H1等5个适体被确定为AFB1的最佳适体。选定的适体用作未修饰的基于金纳米粒子的适体传感器中的探针,以评估它们与 AFB1 的结合亲和力以及它们对其他真菌毒素(黄曲霉毒素 B2、G1、G2、M1、赭曲霉毒素 A 和玉米赤霉烯酮)的选择性。此外,开发了一种新型的直接荧光各向异性适体测定,以确认所选适体在 AFB1 上的结合相互作用。ISM 允许鉴定适体 F20,与亲本适体相比,亲和性和选择性分别提高了 9.4 倍和 2 倍。开发了一种新的直接荧光各向异性适体测定,以确认所选适体在 AFB1 上的结合相互作用。ISM 允许鉴定适体 F20,与亲本适体相比,亲和性和选择性分别提高了 9.4 倍和 2 倍。开发了一种新的直接荧光各向异性适体测定,以确认所选适体在 AFB1 上的结合相互作用。ISM 允许鉴定适体 F20,与亲本适体相比,亲和性和选择性分别提高了 9.4 倍和 2 倍。
更新日期:2020-04-01
中文翻译:

用于生物传感器开发的 DNA 适体对黄曲霉毒素 B1 的亲和力和选择性的计算机成熟
黄曲霉毒素 B1 (AFB1) 的高亲和力和选择性 DNA 适体是通过基于计算机成熟 (ISM) 策略的遗传算法 (GA) 设计的。在许多适体传感器中用作探针的已知 AFB1 适体(专利:PCT/CA2010/001292,Apt1)的序列使用七轮 GA 进行修改,以生成初始文库和三代不同的 ss DNA 寡核苷酸作为新的候选适体。分子对接方法用于筛选和分析最佳适体-AFB1 复合物。此外,还提出了一种新的管道来忠实地预测所有单链 DNA 序列的三级结构。到第二代,适体Apt1序列在局部搜索空间得到优化,F20、g12、C52、C32和H1等5个适体被确定为AFB1的最佳适体。选定的适体用作未修饰的基于金纳米粒子的适体传感器中的探针,以评估它们与 AFB1 的结合亲和力以及它们对其他真菌毒素(黄曲霉毒素 B2、G1、G2、M1、赭曲霉毒素 A 和玉米赤霉烯酮)的选择性。此外,开发了一种新型的直接荧光各向异性适体测定,以确认所选适体在 AFB1 上的结合相互作用。ISM 允许鉴定适体 F20,与亲本适体相比,亲和性和选择性分别提高了 9.4 倍和 2 倍。开发了一种新的直接荧光各向异性适体测定,以确认所选适体在 AFB1 上的结合相互作用。ISM 允许鉴定适体 F20,与亲本适体相比,亲和性和选择性分别提高了 9.4 倍和 2 倍。开发了一种新的直接荧光各向异性适体测定,以确认所选适体在 AFB1 上的结合相互作用。ISM 允许鉴定适体 F20,与亲本适体相比,亲和性和选择性分别提高了 9.4 倍和 2 倍。











































 京公网安备 11010802027423号
京公网安备 11010802027423号