Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Genome-wide association and epistatic interactions of flowering time in soybean cultivar.
PLOS ONE ( IF 2.9 ) Pub Date : 2020-01-22 , DOI: 10.1371/journal.pone.0228114 Kyoung Hyoun Kim 1, 2 , Jae-Yoon Kim 1, 2 , Won-Jun Lim 1, 2 , Seongmun Jeong 1 , Ho-Yeon Lee 1, 2 , Youngbum Cho 1, 2 , Jung-Kyung Moon 3 , Namshin Kim 1, 2
PLOS ONE ( IF 2.9 ) Pub Date : 2020-01-22 , DOI: 10.1371/journal.pone.0228114 Kyoung Hyoun Kim 1, 2 , Jae-Yoon Kim 1, 2 , Won-Jun Lim 1, 2 , Seongmun Jeong 1 , Ho-Yeon Lee 1, 2 , Youngbum Cho 1, 2 , Jung-Kyung Moon 3 , Namshin Kim 1, 2
Affiliation
|
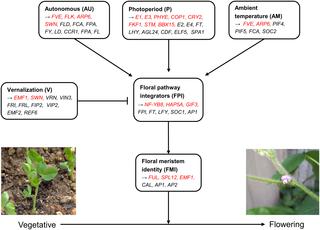
Genome-wide association studies (GWAS) have enabled the discovery of candidate markers that play significant roles in various complex traits in plants. Recently, with increased interest in the search for candidate markers, studies on epistatic interactions between single nucleotide polymorphism (SNP) markers have also increased, thus enabling the identification of more candidate markers along with GWAS on single-variant-additive-effect. Here, we focused on the identification of candidate markers associated with flowering time in soybean (Glycine max). A large population of 2,662 cultivated soybean accessions was genotyped using the 180k Axiom® SoyaSNP array, and the genomic architecture of these accessions was investigated to confirm the population structure. Then, GWAS was conducted to evaluate the association between SNP markers and flowering time. A total of 93 significant SNP markers were detected within 59 significant genes, including E1 and E3, which are the main determinants of flowering time. Based on the GWAS results, multilocus epistatic interactions were examined between the significant and non-significant SNP markers. Two significant and 16 non-significant SNP markers were discovered as candidate markers affecting flowering time via interactions with each other. These 18 candidate SNP markers mapped to 18 candidate genes including E1 and E3, and the 18 candidate genes were involved in six major flowering pathways. Although further biological validation is needed, our results provide additional information on the existing flowering time markers and present another option to marker-assisted breeding programs for regulating flowering time of soybean.
中文翻译:

大豆品种开花时间的全基因组关联和上位相互作用。
全基因组关联研究(GWAS)已经能够发现在植物各种复杂性状中发挥重要作用的候选标记。近年来,随着对候选标记搜索兴趣的增加,对单核苷酸多态性(SNP)标记之间上位相互作用的研究也有所增加,从而能够与单变异加性效应的GWAS一起鉴定更多候选标记。在这里,我们重点鉴定与大豆(Glycine max)开花时间相关的候选标记。使用 180k Axiom® SoyaSNP 阵列对 2,662 个栽培大豆种质进行了基因分型,并对这些种质的基因组结构进行了研究以确认群体结构。然后,进行 GWAS 来评估 SNP 标记与开花时间之间的关联。在59个显着基因中总共检测到93个显着SNP标记,其中E1和E3是开花时间的主要决定因素。根据 GWAS 结果,检查了显着性和非显着性 SNP 标记之间的多位点上位相互作用。发现了两个显着的和 16 个不显着的 SNP 标记作为通过彼此相互作用影响开花时间的候选标记。这18个候选SNP标记映射到E1和E3等18个候选基因,这18个候选基因涉及6个主要的开花途径。尽管需要进一步的生物学验证,但我们的结果提供了有关现有开花时间标记的额外信息,并为调节大豆开花时间的标记辅助育种计划提供了另一种选择。
更新日期:2020-01-23
中文翻译:

大豆品种开花时间的全基因组关联和上位相互作用。
全基因组关联研究(GWAS)已经能够发现在植物各种复杂性状中发挥重要作用的候选标记。近年来,随着对候选标记搜索兴趣的增加,对单核苷酸多态性(SNP)标记之间上位相互作用的研究也有所增加,从而能够与单变异加性效应的GWAS一起鉴定更多候选标记。在这里,我们重点鉴定与大豆(Glycine max)开花时间相关的候选标记。使用 180k Axiom® SoyaSNP 阵列对 2,662 个栽培大豆种质进行了基因分型,并对这些种质的基因组结构进行了研究以确认群体结构。然后,进行 GWAS 来评估 SNP 标记与开花时间之间的关联。在59个显着基因中总共检测到93个显着SNP标记,其中E1和E3是开花时间的主要决定因素。根据 GWAS 结果,检查了显着性和非显着性 SNP 标记之间的多位点上位相互作用。发现了两个显着的和 16 个不显着的 SNP 标记作为通过彼此相互作用影响开花时间的候选标记。这18个候选SNP标记映射到E1和E3等18个候选基因,这18个候选基因涉及6个主要的开花途径。尽管需要进一步的生物学验证,但我们的结果提供了有关现有开花时间标记的额外信息,并为调节大豆开花时间的标记辅助育种计划提供了另一种选择。











































 京公网安备 11010802027423号
京公网安备 11010802027423号