当前位置:
X-MOL 学术
›
J. Biophotonics
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
DeconvTest: Simulation framework for quantifying errors and selecting optimal parameters of image deconvolution.
Journal of Biophotonics ( IF 2.0 ) Pub Date : 2020-02-03 , DOI: 10.1002/jbio.201960079 Anna Medyukhina 1, 2 , Marc Thilo Figge 1, 3
Journal of Biophotonics ( IF 2.0 ) Pub Date : 2020-02-03 , DOI: 10.1002/jbio.201960079 Anna Medyukhina 1, 2 , Marc Thilo Figge 1, 3
Affiliation
|
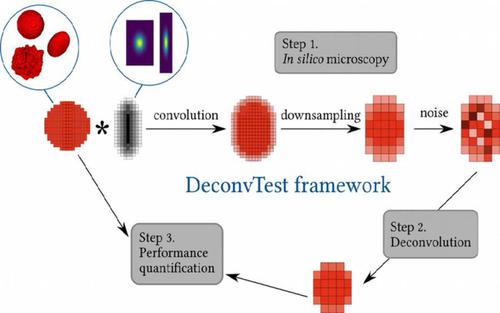
Deconvolution is an essential step of image processing that aims to compensate for the image blur caused by the microscope's point spread function. With many existing deconvolution methods, it is challenging to choose the method and its parameters most appropriate for particular image data at hand. To facilitate this task, we developed DeconvTest: an open‐source Python‐based framework for generating synthetic microscopy images, deconvolving them with different algorithms, and quantifying reconstruction errors. In contrast to existing software, DeconvTest combines all components required to analyze deconvolution performance in a systematic, high‐throughput and quantitative manner. We demonstrate the power of the framework by using it to identify the optimal deconvolution settings for synthetic and real image data. Based on this, we provide a guideline for (a) choosing optimal values of deconvolution parameters for image data at hand and (b) optimizing imaging conditions for best results in combination with subsequent image deconvolution.
中文翻译:

DeconvTest:用于量化误差和选择图像反卷积的最佳参数的仿真框架。
去卷积是图像处理的重要步骤,旨在补偿由显微镜的点扩散功能引起的图像模糊。对于许多现有的反卷积方法,选择最适合手边特定图像数据的方法及其参数具有挑战性。为了简化此任务,我们开发了DeconvTest:这是一个基于Python的开源框架,用于生成合成显微镜图像,使用不同算法对它们进行解卷积并量化重建误差。与现有软件相比,DeconvTest以系统,高通量和定量的方式结合了分析去卷积性能所需的所有组件。我们通过使用它来识别合成和真实图像数据的最佳去卷积设置来展示该框架的功能。基于此,
更新日期:2020-02-03

中文翻译:

DeconvTest:用于量化误差和选择图像反卷积的最佳参数的仿真框架。
去卷积是图像处理的重要步骤,旨在补偿由显微镜的点扩散功能引起的图像模糊。对于许多现有的反卷积方法,选择最适合手边特定图像数据的方法及其参数具有挑战性。为了简化此任务,我们开发了DeconvTest:这是一个基于Python的开源框架,用于生成合成显微镜图像,使用不同算法对它们进行解卷积并量化重建误差。与现有软件相比,DeconvTest以系统,高通量和定量的方式结合了分析去卷积性能所需的所有组件。我们通过使用它来识别合成和真实图像数据的最佳去卷积设置来展示该框架的功能。基于此,












































 京公网安备 11010802027423号
京公网安备 11010802027423号