Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Antimicrobial resistance patterns and molecular resistance markers of Campylobacter jejuni isolates from human diarrheal cases.
PLOS ONE ( IF 2.9 ) Pub Date : 2020-01-17 , DOI: 10.1371/journal.pone.0227833 Mohamed Elhadidy 1, 2 , Mohamed Medhat Ali 1, 3 , Ayman El-Shibiny 1, 4 , William G Miller 5 , Walid F Elkhatib 6, 7 , Nadine Botteldoorn 8 , Katelijne Dierick 9
PLOS ONE ( IF 2.9 ) Pub Date : 2020-01-17 , DOI: 10.1371/journal.pone.0227833 Mohamed Elhadidy 1, 2 , Mohamed Medhat Ali 1, 3 , Ayman El-Shibiny 1, 4 , William G Miller 5 , Walid F Elkhatib 6, 7 , Nadine Botteldoorn 8 , Katelijne Dierick 9
Affiliation
|
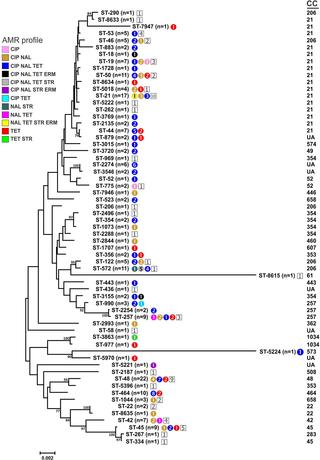
The aim of this study is to characterize the antimicrobial resistance of Campylobacter jejuni recovered from diarrheal patients in Belgium, focusing on the genetic diversity of resistant strains and underlying molecular mechanisms of resistance among Campylobacter jejuni resistant strains isolated from diarrheal patients in Belgium. Susceptibility profile of 199 clinical C. jejuni isolates was determined by minimum inhibitory concentrations against six commonly-used antibiotics (ciprofloxacin, nalidixic acid, tetracycline, streptomycin, gentamicin, and erythromycin). High rates of resistance were observed against nalidixic acid (56.3%), ciprofloxacin (55.8%) and tetracycline (49.7%); these rates were similar to those obtained from different national reports in broilers intended for human consumption. Alternatively, lower resistance rates to streptomycin (4.5%) and erythromycin (2%), and absolute sensitivity to gentamicin were observed. C. jejuni isolates resistant to tetracycline or quinolones (ciprofloxacin and/or nalidixic acid) were screened for the presence of the tetO gene and the C257T mutation in the quinolone resistance determining region (QRDR) of the gyrase gene gyrA, respectively. Interestingly, some of the isolates that displayed phenotypic resistance to these antimicrobials lacked the corresponding genetic determinants. Among erythromycin-resistant isolates, a diverse array of potential molecular resistance mechanisms was investigated, including the presence of ermB and mutations in the 23S rRNA gene, the rplD and rplV ribosomal genes, and the regulatory region of the cmeABC operon. Two of the four erythromycin-resistant isolates harboured the A2075G transition mutation in the 23S rRNA gene; one of these isolates exhibited further mutations in rplD, rplV and in the cmeABC regulatory region. This study expands the current understanding of how different genetic determinants and particular clones shape the epidemiology of antimicrobial resistance in C. jejuni in Belgium. It also reveals many questions in need of further investigation, such as the role of other undetermined molecular mechanisms that may potentially contribute to the antimicrobial resistance of Campylobacter.
中文翻译:

从人类腹泻病例中分离出的空肠弯曲菌的抗菌素耐药性模式和分子耐药性标记。
本研究的目的是表征从比利时腹泻患者中回收的空肠弯曲菌的抗菌药物耐药性,重点关注从比利时腹泻患者中分离出的空肠弯曲菌耐药菌株的遗传多样性和潜在的耐药分子机制。通过对六种常用抗生素(环丙沙星、萘啶酸、四环素、链霉素、庆大霉素和红霉素)的最低抑制浓度确定了 199 个临床空肠弯曲菌分离株的敏感性。对萘啶酸(56.3%)、环丙沙星(55.8%)和四环素(49.7%)的耐药率很高;这些比率与从供人类消费的肉鸡的不同国家报告中获得的比率相似。另外,观察到对链霉素(4.5%)和红霉素(2%)的耐药率较低,对庆大霉素的绝对敏感性也较低。分别筛选对四环素或喹诺酮类药物(环丙沙星和/或萘啶酸)耐药的空肠弯曲菌分离株,以确定是否存在tetO基因以及促旋酶基因gyrA的喹诺酮耐药决定区(QRDR)中是否存在C257T突变。有趣的是,一些对这些抗菌药物表现出表型耐药性的分离株缺乏相应的遗传决定因素。在红霉素耐药菌株中,研究了多种潜在的分子耐药机制,包括 ermB 的存在以及 23S rRNA 基因、rplD 和 rplV 核糖体基因以及 cmeABC 操纵子的调控区域的突变。 四种红霉素抗性分离株中的两种在 23S rRNA 基因中含有 A2075G 过渡突变;其中一种分离株在 rplD、rplV 和 cmeABC 调控区中表现出进一步的突变。这项研究扩展了目前对不同遗传决定因素和特定克隆如何影响比利时空肠弯曲菌抗生素耐药性流行病学的理解。它还揭示了许多需要进一步研究的问题,例如可能导致弯曲杆菌抗菌素耐药性的其他未确定的分子机制的作用。
更新日期:2020-01-21
中文翻译:

从人类腹泻病例中分离出的空肠弯曲菌的抗菌素耐药性模式和分子耐药性标记。
本研究的目的是表征从比利时腹泻患者中回收的空肠弯曲菌的抗菌药物耐药性,重点关注从比利时腹泻患者中分离出的空肠弯曲菌耐药菌株的遗传多样性和潜在的耐药分子机制。通过对六种常用抗生素(环丙沙星、萘啶酸、四环素、链霉素、庆大霉素和红霉素)的最低抑制浓度确定了 199 个临床空肠弯曲菌分离株的敏感性。对萘啶酸(56.3%)、环丙沙星(55.8%)和四环素(49.7%)的耐药率很高;这些比率与从供人类消费的肉鸡的不同国家报告中获得的比率相似。另外,观察到对链霉素(4.5%)和红霉素(2%)的耐药率较低,对庆大霉素的绝对敏感性也较低。分别筛选对四环素或喹诺酮类药物(环丙沙星和/或萘啶酸)耐药的空肠弯曲菌分离株,以确定是否存在tetO基因以及促旋酶基因gyrA的喹诺酮耐药决定区(QRDR)中是否存在C257T突变。有趣的是,一些对这些抗菌药物表现出表型耐药性的分离株缺乏相应的遗传决定因素。在红霉素耐药菌株中,研究了多种潜在的分子耐药机制,包括 ermB 的存在以及 23S rRNA 基因、rplD 和 rplV 核糖体基因以及 cmeABC 操纵子的调控区域的突变。 四种红霉素抗性分离株中的两种在 23S rRNA 基因中含有 A2075G 过渡突变;其中一种分离株在 rplD、rplV 和 cmeABC 调控区中表现出进一步的突变。这项研究扩展了目前对不同遗传决定因素和特定克隆如何影响比利时空肠弯曲菌抗生素耐药性流行病学的理解。它还揭示了许多需要进一步研究的问题,例如可能导致弯曲杆菌抗菌素耐药性的其他未确定的分子机制的作用。









































 京公网安备 11010802027423号
京公网安备 11010802027423号